1000
An Unsupervised Deep Learning Method for Parallel MR Cardiac Imaging via Time Interleaved Sampling1Research Center for Medical AI, Shenzhen Institutes of Advanced Technology,Chinese Academy of Sciences, Shenzhen, China, 2Shenzhen College of Advanced Technology, University of Chinese Academy of Sciences, Shenzhen, China, 3Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institutes of Advanced Technology,Chinese Academy of Sciences, Shenzhen, China, 4Department of Biomedical Engineering and Department of Electrical Engineering, The State University of New York, Buffalo, NY, United States
Synopsis
Deep learning has achieved good success in cardiac MRI. However, these methods are all based on big data, and only deal with single-channel imaging. In this paper, we propose an unsupervised deep learning method for parallel MR cardiac imaging via time interleaved sampling. Specifically, a set of full-encoded reference data were built by merging the data from adjacent time frames, and used to train a network for reconstructing each coil image separately. Finally, coil images were combined via another CNN. The validation on in vivo data show that our method can achieve improved reconstruction compared with other competing methods.
Introduction
It is important to improve the temporal and spatial resolution of cardiac MR imaging by using fast imaging method. Although the traditional optimization methods [1-3] based on sparse or low rank have achieved good success, some issues still remain such as difficult to choose the hyper-parameters, long iteration time and losing details. Recently, deep learning has been introduced to speed up dynamic MR reconstruction. For example, DC-CNN [4] uses a deep cascade of CNNs to accelerate the data acquisition process by combining data consistency and data sharing approaches. In CRNN [5], spatio-temporal dependencies are simultaneously learnt by exploiting bidirectional recurrent hidden connections across time sequences. DIMENSION [6] developed a multi-supervised network training technique to constrain frequency domain and spatial domain information simultaneously. Although these works make great contributions to dynamic MR imaging, the following issues still need to be addressed: 1) all these methods require a large amount of full-sampled cardiac MR images. However, the collections of full-sampled k-space data are always difficult, especially breath-holding are needed in the acquisition scheme; 2) the inputs of these methods are all single-coil signals, which did not use the information between coils as well as not accord with the principle of parallel magnetic resonance acquisition. Our work mainly focuses on solving these two issues.Theory and method
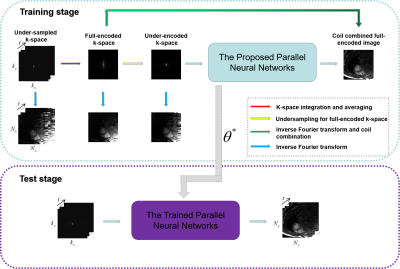
In this paper, we propose an unsupervised deep learning method for parallel MR cardiac imaging via time interleaved sampling. The diagram of the proposed framework is shown in Figure 1. In particular, we designed a time interleaved acquisition scheme to build a set of full-encoded reference data by merging the adjacent time frames. Then these full-encoded data can be used to train a coil-by-coil parallel network to reconstruct the image of each coil separately. Finally, the coil images were combined together via another convolutional neural network.The whole unsupervised framework can be divided into three components: 1) Data preparation: A representative acquisition scheme for R = 5 is shown in Figure 2. The acquired data from adjacent time frames can be merged to build a complete set of k-space data. Once the full-encoded data set is built, the data pairs of network input and output can be obtained by retrospective under-sampling the full-encoded data. Although the training data sets are synthetic, they can effectively represent true sampled data and eliminate temporal redundancy. 2) Network training: For parallel reconstruction of under-encoded multi-coil k-space data, we propose a novel parallel network in a coil-by-coil manner shown in Figure 3. For each coil, there is a separate network to reconstruct it. We selected ADMM-Net-III [7] as the reconstruction network. Then all of these reconstructed coil images are concatenated together to fed into another CNNs to explore coil correlation. 3) Online test: The scanned in vivo under-sampled k-space data is fed into the trained network to reconstruct 2D dynamic MR images. This unsupervised framework has many advantages: no need for full-sampled dynamic MR data set, coil correlation can be explored, no need for coil sensitivity maps, and elimination of time redundancy.
Experiment and results
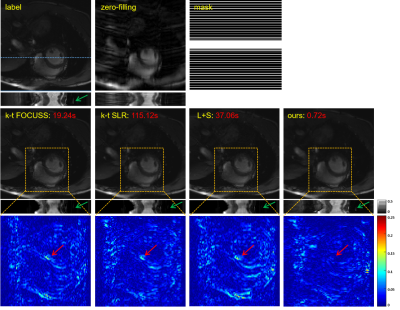
We collected 386 2DT (refer to 2D dynamic) full-sampled cardiac MR data from 30 healthy volunteers using 3T scanner (SIEMENS MAGNETOM Trio) with T1-weghted bSSFP sequence. After normalization and extraction, we got 2149 cardiac data for network training. For each of these, we retrospectively under-sampled it with 16 ACS lines according to the time-interleaved scheme. The models were implemented on an Ubuntu 16.04 LTS (64-bit) operating system equipped with an Intel Xeon E5-2640 (CPU) and a Tesla TITAN Xp (GPU, 12GB memory). The open framework Tensorflow was used.To demonstrate the efficacy of the proposed unsupervised learning method, we compared with k-t FOCUSS, k-t SLR, and L+S. Specifically, we adjusted the parameters of the CS-MRI methods to their best performance. The reconstructions of these methods are shown in Figure 4. The three CS-based reconstruction results contain less structural details and more artifacts than that of the proposed method. We also enlarge the error maps of the cardiac region, which show that our method has the best reconstruction performance on the cardiac region, especially the details marked by the red arrow. A possible reason that the artifacts are not completely removed in the reconstructions of three CS-based methods is that fewer frames were collected and a uniform sampling pattern was used, which they may be not good at. The y-t images, which are extracted from 124-th slice along the y and temporal dimensions, can also clearly illustrate the superior performance of the proposed method. As indicated by the green arrow, only the proposed method has accurately reconstructed this vessel. The red numbers in the second row represent reconstruction time of these methods and our method is hundreds of times shorter than the competing methods.
Conclusion
In this paper, we propose an unsupervised deep learning method for parallel MR cardiac imaging via time interleaved sampling. The comparisons with classical k‐t FOCUSS, k‐t SLR, L+S on in vivo data show our method can achieve improved reconstruction results. This unsupervised framework has many advantages: no need for full-sampled dynamic MR data set, coil correlation can be explored, no need for coil sensitivity maps, elimination of time redundancy.Acknowledgements
Grant support: China NSFC 61601450, 61871371, Science and Technology Planning Project of Guangdong Province (2017B020227012, 2018B010109009), the Basic ResearchProgram of Shenzhen (JCYJ20180507182400762, JCYJ20150831154213680), Youth Innovation Promotion Association Program of ChineseAcademy of Sciences (2019351), and the Innovation and Technology Commission of the government of Hong Kong SAR (MRP/001/18X).References
[1]. Jung, Hong, Jong Chul Ye, and Eung Yeop Kim. "Improved k–t BLAST and k–t SENSE using FOCUSS." Physics in Medicine & Biology 52, no. 11 (2007): 3201.
[2]. Lingala, Sajan Goud, Yue Hu, Edward DiBella, and Mathews Jacob. "Accelerated dynamic MRI exploiting sparsity and low-rank structure: kt SLR." IEEE transactions on medical imaging30, no. 5 (2011): 1042-1054.
[3]. Otazo, Ricardo, Emmanuel Candès, and Daniel K. Sodickson. "Low‐rank plus sparse matrix decomposition for accelerated dynamic MRI with separation of background and dynamic components." Magnetic Resonance in Medicine 73, no. 3 (2015): 1125-1136.
[4]. J. Schlemper, J. Caballero, J.V. Hajnal, A. Price, D. Rueckert, “A Deep Cascade of Convolutional Neural Networks for Dynamic MR Image Reconstruction”, IEEE TMI, DOI: 10. 1109/TMI.2017.2760978 (2017)
[5]. Qin, Chen, Joseph V. Hajnal, Daniel Rueckert, Jo Schlemper, Jose Caballero, and Anthony N. Price. "Convolutional recurrent neural networks for dynamic MR image reconstruction." IEEE transactions on medical imaging (2018).
[6]. Wang S, Ke Z, Cheng H, et al. DIMENSION: Dynamic MR imaging with both k-space and spatial prior knowledge obtained via multi-supervised network training. NMR in Biomedicine. 2019;e4131. https://doi.org/10.1002/nbm.4131.
[7]. Cheng J, Wang H, Zhu Y, Liu Q, Ying L, Liang D. Model-based Deep MR Imaging: the roadmap of generalizing compressed sensing model using deep learning. arXiv preprint arXiv:1906.08143. 2019 Jun 19.
Figures