0999
Dynamic Real-time MRI with Deep Convolutional Recurrent Neural Networks and Non-Cartesian Fidelity1Institute for Medical Imaging Technology, School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China, 2United Imaging Healthcare, Shanghai, China
Synopsis
A convolutional recurrent neural networks (CRNN) with Non-Cartesian fidelity for 2D real-time imaging was proposed. 3D stack-of-star GRE radial sequence with self-navigator was used to acquire the data. Multiple respiratory phases were extracted from the navigator and the sliding window method was used to get the training data. The Fidelity constraints the reconstruction image to be consistent to the undersampled non-Cartesian k-space data. Convolution and recurrence improve the quality of the reconstructed images by using temporal dimension information. The reconstruction speed is around 10 frames/second, which fulfills the requirement of real-time imaging.
Introduction
Real-time MRI would be helpful in MR-guided surgery, medical robot, intervention, etc1. In interventional surgery, the electrode is sometimes inserted into the wrong place of the body due to patient movement or incorrect registration during surgery preparation. Real-time imaging would be useful for physicians to avoid this unfavorable situation, and improve the patient outcome. MRI is intrinsically slow due to the physical and physiological limitation. With the successful development of compressed sensing (CS) theory2,3, several CS techniques have been applied to accelerate imaging4,5. However, CS based methods are computational expensive, and is difficult for real-time imaging. Deep learning gives a new opportunity, due to its fast and high-quality reconstruction. While most of the deep learning methods is designed for Cartesian sampled, Cartesian is not as good as non-Cartesian sampled for dynamic MRI due to its limitation in k-space traverse. Here, we extend the deep convolutional recurrent neural network (CRNN)6, and combine with the non-Cartesian Fidelity module for real-time dynamic MRI. Trained with 3D stack-of-star GRE radial data, the Non-Cartesian CRNN (NCRNN) network reconstructs the 2D GRE radial data with a temporal resolution of 100ms and fulfils the requirements of real-time imaging.Theory and Methods
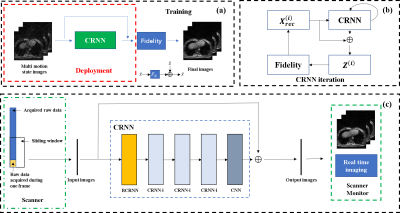
In real-time imaging application, the NCRNN network learns the correlations in spatial and temporal domain, and learns the correlations among iterations. As shown in Fig. 1 (c), the input of the initial iteration is composed of undersampled images for different respiratory phases, and the target of the output is composed of fully sampled images. The Fidelity constraints the reconstruction image to be consistent to the undersampled k-space data. The output of the i-th iteration is inputted to the next iteration, which will improve the quality of the output over iterations. The Fidelity is computed as $$$ \hat{X}=\tilde{X}+\alpha {{F}^{-1}}(Y-\beta MF\tilde{X})$$$, where $$$\tilde{X}$$$ is the output of the CNN layers in CRNN, $$$F$$$ is NUFFT operator and $$${F}^{-1}$$$ is inversion NUFFT operator, $$$M$$$ is the undersampling mask. $$$Y$$$ is undersampled k-space data, $$$\alpha$$$ is trainable parameters for regularization, $$$\beta$$$ learns the scaling factor of NUFFT operator, $$$\hat{X}$$$ is the output of the Fidelity. All the data were acquired on a 3T MRI scanner (uMR790, United Imaging Healthcare, Shanghai, China), by 3D stack-of-star GRE radial sequence with self-navigator. The center of radial k-space line was used for navigator. Thirty respiratory phases were divided by PCA method, and the ground truth images are reconstructed by XD-GRASP7. Thorax and abdomen images were undersampled using reduction factor R=4. The training data has 7200 images in total (8 volunteers×30 slices×30 respiratory phases). 5400 images were used for training, 900 images were used for validation and 900 images were used for test. The 4X undersampled aliased images were inputted to NCRNN, and fully sampled images were used as ground truth. During the training, the input includes 30 images for different respiratory phases (matrix size is 128×128×30). While during the test, the input includes 30 and 10 images for different respiratory phases images (matrix size is 128×128×30 and 128×128×10). The NCRNN was implemented in Pytorch8.Results & Discussion
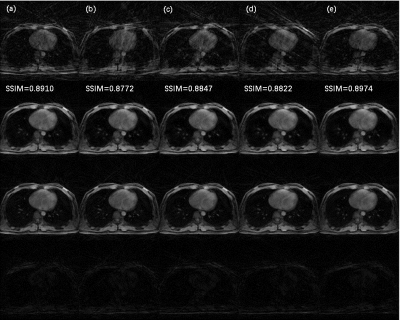
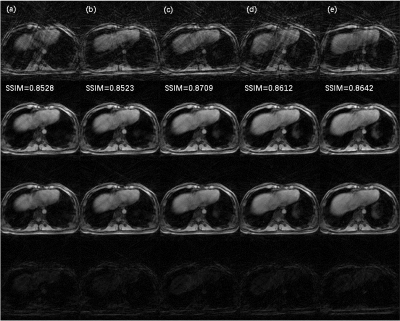
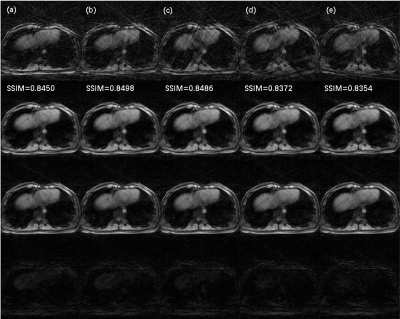
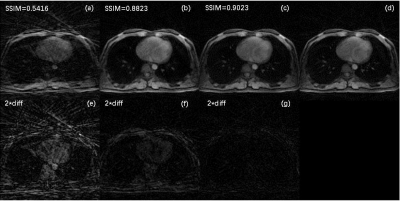
The architecture of NCRNN and overall procedure of real-time imaging method by Gadgetron9 were shown in Fig. 1. Fig.2-4 show the reconstructed results using NCRNN. In Fig.2-4, Columns (a-e) represent images for different respiratory phases. The first row is the 4X undersampled images, which are the input of the NCRNN network. The second row is the output of the NCRNN network for each respiratory phase. The third row is the fully sampled images, which are the ground truth during the NCRNN training. The fourth row is the difference between the ground truth and the output for each respiratory phase. In Fig. 2 and Fig. 3 show reconstructed results of two different slice, and the input has 30 different respiratory phases. Fig.4 shows the reconstructed results of the same slice in Fig.3, but the input has 10 different respiratory phases. The NCRNN show the compatibility for different input size. Fig. 5 shows the comparison between NCRNN and XD-GRASP. (a) NUFFT reconstructed 4X undersampled image. (b) Output of the NCRNN network, with structural similarity index (SSIM) = 0.8823 compared with the ground truth. (c) XD-GRASP reconstruction, with SSIM = 0.9023 compared with the ground truth. (d) The ground truth. The absolute value of difference times 2 of the fully sampled image with reconstrued images of different methods were shown in (e-g).Conclusion
We proposed and evaluated a deep learning reconstruction method for real-time imaging by NCRNN network. The in vivo results demonstrate that at acceleration rate R=4, the reconstruction quality of NCRNN is comparable to XD-GRASP, while the reconstruction time is only 100ms compared with 40s of XD-GRASP, fulfilling the requirements of real-time imaging.Acknowledgements
The authors thank United Imaging Healthcare for making the United Imaging Healthcare(UIH) Application Development Environment and Programming Tools (ADEPT) available for convenient development of the MRI pulse sequences and real-time imaging system.References
[1] Emmanuel Candes, et al. Robust uncertainty principles: exact signal reconstruction from highly incomplete frequency information. IEEE Trans Inf Theory. 2006; 52:489‐509.
[2] Donoho DL. Compressed sensing. IEEE Trans Inf Theory. 2006; 52:1289‐1306.
[3] Lingala SG, et al. Blind compressed sensing dynamic MRI. IEEE Trans Med Imaging. 2013; 32:1132‐1145.
[4] Otazo R, et al. Combination of Compressed Sensing and Parallel Imaging for Highly Accelerated First-Pass Cardiac Perfusion MRI. MRM 2010; 64:767–776.
[5] Lustig M, et al. Sparse MRI: The Application of Compressed Sensing for Rapid MR Imaging. MRM 2007; 58:1182–1195.
[6] Qin C, et al. Convolutional Recurrent Neural Networks for Dynamic MR Image Reconstruction. arXiv:1712.01751.
[7] Feng, et al. XD‐GRASP: Golden‐angle radial MRI with reconstruction of extra motion‐state dimensions using compressed sensing. MRM 2016; 75:775–788.
[8] https://pytorch.org
[9] http://gadgetron.github.io
Figures