0986
Sliding-Slab Profile Encoding (SLIPEN) for Eliminating Slab Boundary Artifact in Three-Dimensional Multi-slab Diffusion-Tensor Imaging1Department of Diagnostic Radiology, The University of Hong Kong, Hong Kong, China, 2Department of Radiology and Biomedical Imaging, University of California San Francisco, San Francisco, CA, United States, 3The State Key Laboratory of Brain and Cognitive Sciences, Hong Kong, China, 4Philips Healthcare, Hong Kong, China
Synopsis
3D multi-slab diffusion-tensor imaging (DTI) can enable high-resolution DTI at submillimeter voxel size. However, the slab boundary artifact and distortion along slab direction can deteriorate the data quality of 3D DTI, thereby limiting its applications. In this work, we proposed a sliding-slab profile encoding (SLIPEN) method to acquire the 3D multi-slab DTI data with sliding-slab technique, and to reconstruct the data free from slab boundary artifact. In addition, off-resonance correction can be incorporated into SLIPEN for producing high-quality artifact-free 3D DTI data.
Purpose
3D multi-slab diffusion-tensor imaging can achieve higher spatial resolution with improved SNR efficiency, therefore being superior to 2D DTI [1]. However, because of non-ideal excitation profile and crosstalk effect, the multi-slab acquisition often leads to slab boundary artifact (SBA) [2,3,4]. Moreover, the off-resonance effect can cause distorted slab profile during excitation, leading to aliasing artifact along slice-encoding direction in the final reconstruction images [6]. Thus, additional correction is required to produce high-quality 3D DTI data. In this work, we proposed to address aforementioned issues associated with multi-slab acquisition using sliding-slab profile encoding (SLIPEN) method, by incorporating additional slab-profile encoding and off-resonance correction into the reconstruction framework.Methods
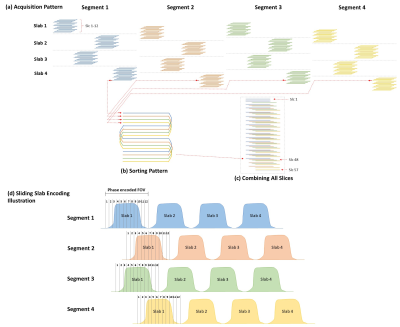
Acquisition Strategy: We have enabled sliding-slab technique [6,7] for acquiring different in-plane segments of 3D multi-slab DTI data with interleaved-EPI readout. To avoid spin history effect, the acquisition of in-plane segment is arranged at outermost loop of entire data acquisition (Fig.1a). Afterward, all in-plane segment data acquired with different slice(kz) encoding from multiple slabs are sorted by slice location, to generate the full 3D image volume (Figs.1b&c). To minimize the crosstalk effect between two adjacent slabs, a gap with 50% of effective slab thickness is used (Fig.1d). The interleaved sliding-slab pattern is also applied to improve the performance of following reconstruction.Data Reconstruction: The proposed SLIPEN reconstruction method mainly consists of three reconstruction stages, summarized in Fig.2. The preprocessing steps including noise calibration, Nyquist ghost correction, and measurement of inter-shot phase variations are similar as 3D-MUSE method [4]. An additional field map data is acquired to estimate the phase accumulation $$$\Phi_{j}$$$ (for j-th kz encoding) for slab distortion correction.
In the first stage, the direct reconstruction volume image indexed by m, is reconstructed from each segment data $$$d_{m,j,k}$$$ (k represents k-th multi-coil) separately by solving Eq[2] with CG method [9].
$$X_{m} = argmin\left \{ \left \| exp(i\Phi _{j})FC_{k}exp(i\varphi _{m,j})X_{m}-d_{m,j,k} \right \|_{2}^{2} \right \} \qquad[1]$$
where $$$F$$$ represents the Fourier transform, $$$C_{k}$$$ the coil sensitivity, and $$$\varphi_{m,j}$$$ the inter-shot phase variation. Afterward, we use two sliding-summation windows with different kernel sizes [10] to measure the weighting function ($$$W_{m}$$$) of slab profiles for m-th segment data and combine all $$$X_{m}$$$ with weightings to generate the joint reconstruction volume image $$$X$$$.
In the second stage, the high quality joint reconstruction image volume and accurate slab profiles were jointly estimated via an iterative reconstruction algorithm with results from the first stage as initial input. The problem can be reformulated as an optimization problem.
$$\left \{ X,W_{m} \right \} = argmin\left \{ \left \| exp(i\Phi _{j})FC_{k}exp(i\varphi _{m,j})W_{m}X-d_{m,j,k} \right \|_{2}^{2}+\lambda \left \| W_{m}-\overline{W} \right \|_{2}^{2} \right \} \qquad[2]$$
where is the final artifact-free image volume, other notations are kept the same as in Eq[1]. An additional penalty on $$$W$$$ is added to improve the model robustness. $$$\overline{W}$$$ represents the mean of $$$W_{m}$$$, and $$$\lambda$$$ the penalty weighting. The $$$X$$$ and $$$W_{m}$$$ are solved by using alternative gradient descent method.
In-vivo Experiment: Human brain 3D DTI data with 1.3-mm isotropic resolution was acquired from a 3.0T MRI (Philips, Achieva) using proposed acquisition strategy with 4 in-plane segments and 32-channel head coil. Scan parameters: FOV = 208x208x152mm3, number of slabs = 9, kz encoding per slab = 12, effective slab thickness (for RF) = 10.4mm, RF bandwidth = 565Hz, TE1/TE2/TR = 61/112/2000ms, PF factor = 0.71, number of DTI = 6, b = 800s/mm2, and scan time = 11.3min.
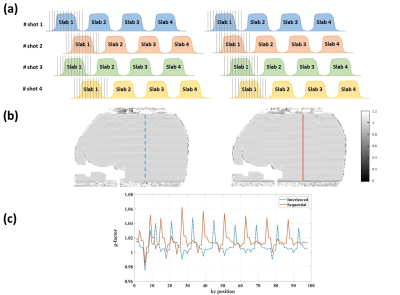
Evaluation: We compared different sliding-slab acquisition patterns to evaluate the reconstruction performance of SLIPEN method by calculating g-factor maps and comparing DTI image quality.
Results
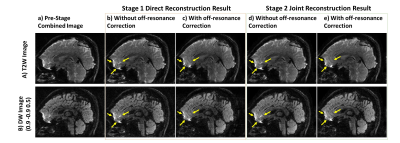
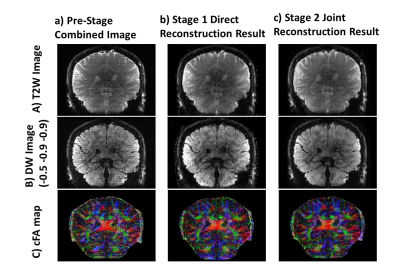
Fig.3&Fig.4 show the reconstructed data by SLIPEN framework at different stages in sagittal and coronal view, respectively. At pre-stage, shown in Fig.3(a)&Fig.4(a), the images suffer from severe SBA and off-resonance effect when directly combining multi-slab data. At first stage, shown in Fig.3(b,c)&Fig.4(b), the SBA can be largely eliminated, but the region with strong off-resonance effect, pointed by yellow arrows in Fig.3(b), still has aliasing artifacts. At the same time, the bright spots near the edges of skull, shown in Fig.4(b), indicate incomplete convergence at first stage. At second stage, shown in Fig.3(d,e)&Fig.4(c), the iterative reconstruction framework can suppress the noise in the middle of joint reconstructed volume data and improve image quality. Fig.5 evaluates two different sliding-slab acquisition patterns in Fig.5(a) by calculating g-factor maps in Fig.5(b). A line profile in Fig.5(c) is chosen for sharpness comparison. Compared to sequential sliding-slab acquisition, interleaved sliding-slab pattern has little effect on g-factor map.Discussion and Conclusion
In this work, we demonstrated that the proposed SLIPEN method with iterative correction algorithms can effectively produce high-quality 3D DTI data without SBA and distortion along slab direction. A major advantage of SLIPEN is no need to acquire additional data or measure slab profile prior to acquisition of imaging data. Moreover, the proposed joint reconstruction of combined image volume (stage2) and slab profiles outperforms the standard slab profiles estimation method (stage 1). In our preliminary evaluation, the interleaved sliding-slab pattern has little effect on g-factor maps. Further investigation is required to optimize the SNR efficiency associated with sliding-slab pattern. In conclusion, SLIPEN can eliminate SBA in 3D multi-slab DTI, therefore making 3D DTI more practical for neuroscience research.Acknowledgements
The work was in part supported by grants from Hong Kong Research Grant Council (GRF HKU17138616 and GRF HKU17121517), and Hong Kong Innovation and Technology Commission (ITS/403/18).References
1. Frost R., Miller K.L., Tijssen R.H.N., Porter D.A., Jezzard P. 3D Multi‐slab diffusion‐weighted readout‐segmented EPI with real‐time cardiac‐reordered k‐space acquisition. MRM 2014; 72:1565-1579.
2. Van A.T., Aksoy M., Holdsworth S.J., Kopeinigg D., Vos S.B., Bammer R. Slab Profile Encoding (PEN) for Minimizing Slab Boundary Artifact in Three-Dimensional Diffusion-Weighted Multislab Acquisition. MRM 2015; 73:605–613.
3. Wu W., Koopmans P.J., Frost R., Miller K.L. Reducing Slab Boundary Artifacts in Three-Dimensional Multislab Diffusion MRI Using Nonlinear Inversion for Slab Profile Encoding (NPEN). MRM 2016; 76:1183-1195.
4. Chang H.C., Hui E.S., Chiu P.W., Liu X., Chen N.K. Phase Correction for Three-Dimensional (3D) Diffusion-Weighted Interleaved EPI Using 3D Multiplexed Sensitivity Encoding and Reconstruction (3D-MUSER). MRM 2018; 79:2702-2712.
5. Lu W., Pauly K.B., Gold G.E., Pauly J.M., Hargreaves B.A. SEMAC: Slice Encoding for Metal Artifact Correction in MRI. MRM 2009; 62:66-76.
6. Li Z., Wang D., Robison R.K., Zwart N.R., Schar M., Karis J.P., Pipe J.P. Sliding-Slab Three-Dimensional TSE Imaging With a Spiral-In/Out Readout. MRM 2016; 75:729-738.
7. Liu K., Xu Y., Loncar M. Reduced Slab Boundary Artifact in Multi-slab 3D Fast Spin-Echo Imaging. MRM 2000; 44:269–276.
8. Chen N.K., Avram A.V., Song A.W. Two-Dimensional Phase Cycled Reconstruction for Inherent Correction of Echo-Planar Imaging Nyquist Artifacts. MRM 2011; 66:1057–1066.
9. Liu X., Cui D., Dai E., Hui E.S., Chan Q., Chang H.C. Self-calibrated and Collaborative Propeller-EPI Reconstruction (SCOPER) for High-Quality Diffusion-Tensor Imaging. In Proceedings of the Joint Annual Meeting of ISMRM-ESMRMB, Montreal, Canada, 2019. Abstract 230.
10. Man L.C., Pauly J.M., Macovski A. Improved Automatic Off-Resonance Correction Without a Field Map in Spiral Imaging. MRM 1997; 37:906-913.
Figures