0984
Structure preserving noise removal in Hilbert space from ultra-high resolution diffusion MRI data1Brigham and Women's Hospital, Harvard Medical School, Boston, MA, United States, 2Massachusetts General Hospital, Harvard Medical School, Charlestown, MA, United States
Synopsis
In-vivo submillimeter resolution diffusion MRI suffers from limited signal-to-noise ratio (SNR) due to the small voxel size. Denoising techniques can improve the SNR and facilitate further dMRI analysis. Among them, perhaps PCA-based (e.g, Marchenko-Pastur PCA) have shown the best performance. In this work, we introduce kernel PCA, a powerful nonlinear generalization of linear PCA to Hilbert spaces that is shown to suppress a substantial amount of noise (which MP-PCA is incapable of) and still reliably preserve dMRI signal. We showcase K-PCA noise removal with 660 micrometer gSlider data, where we compared it qualitatively and qualitatively with MP-PCA.
Introduction
Ultra-high resolution diffusion MRI holds the promise of being an essential tool to explore human brain at an unprecedented level of detail. However, at that resolution, severe noise often obscures fine anatomical details and hampers further dMRI analysis. We introduce kernel PCA (principal component analysis), a non-linear generalization of the traditional linear PCA to Hilbert spaces, to suppress noise while preserving the fine details in the diffusion-weighted images. The proposed method holds promise for denoising at very low SNR levels outperforming standard PCA denoising methods.Methods
Kernel PCAKernel PCA (K-PCA) is a nonlinear extension of the traditional linear PCA where principal component analysis is performed not in the native space, but in a high dimensional reproducing kernel Hilbert space (RKHS) where the data lies on a flat manifold1. Let the input data set be $$$\{ {\bf{x}}_k \in \mathbb{R}^D, k=1,...,N\}$$$ and the mapped data be $$$\psi({\bf{x}}_k ) \in F$$$, where $$$\psi(\cdot)$$$ is a non-linear mapping from the original space to the RKHS1,2. As in PCA, the directions of maximum variance in the RKHS feature space can be obtained using singular value decomposition in the feature space $$$F$$$1,2. The power of K-PCA lies in its ability to capture non-linear dependencies/redundancies that may exist in the data, i.e., diffusion signal, and that are not possible to model with linear PCA. In terms of denoising, given an input vector (dMRI signal), $$$ \bf x $$$, a denoised version $$$\hat{\bf x }$$$ is obtained as a pre-image of the projection $$$P\psi( \bf x )$$$ of $$$ \bf x $$$ in the feature space. To accomplish the whole process, there is no need to know the mapping $$$\psi(\cdot)$$$ explicitly but just to define a kernel function, $$$K( {\bf{x}}_k , {\bf{x}}_{k'})$$$, which measures the similarity between two vectors $$${\bf{x}}_k$$$ and $$${\bf{x}}_{k'}$$$,3 allowing to define a metric on $$$F$$$1,2.
Experiments
We demonstrate that K-PCA denoising outperforms state-of-the-art linear PCA based denoising methods, e.g. Marchenko-Pastur PCA, MP-PCA4,5, in removing noise while preserving the fine structure in ultra-high resolution dMRI data. We implement K-PCA in a voxel-wise fashion with a patch size of [5x5x5]. A Gaussian kernel is a typical choice for KPCA with bandwith $$$\sigma$$$ defined automatically and adaptively in a data-driven manner as given in6-8. To obtain the denoised signal $$$\hat{\bf x }$$$ in closed-form, we used the method presented in8. We note that no truncation of the eigenvalues is required in this method, unlike MP-PCA. Finally, MP-PCA was implemented in a similar voxel-wise fashion with the same patch size for comparative analysis.
Dataset:
Whole-brain gSlider-SMS dMRI data (D = 64 directions, $$$b = 1500 s/{mm}^2$$$, 7b0 volumes) was acquired (sagittally), with three repetitions, and reconstructed with conventional gSlider processing9 to create 660 $$${\mu}m$$$ isotropic resolution data( gslider-1av). K-PCA and MP-PCA were then used to denoise the 64 images of one of the three DWI datasets (gslider-1av) and compared to a reference: gSlider reconstruction averaged three times(gSlider-3av).
Validation:
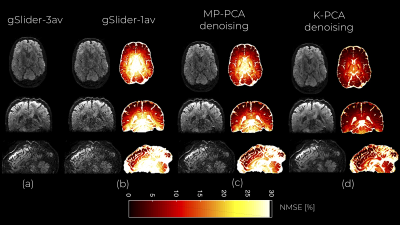
We used the normalized mean square error (NMSE) of the DWI signals and the relative absolute error of the fractional anisotropy (FA), and mean diffusivity (MD) estimated after applying DTI (dtifit FSL) to compare the denoised DWI datasets. Reference FA and MD values were those derived from gSlider-3av: $$$FA_{3AV}$$$, $$$MD_{3AV}$$$ (estimated with the same DTI algorithm).
Results
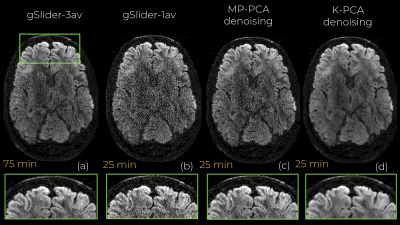
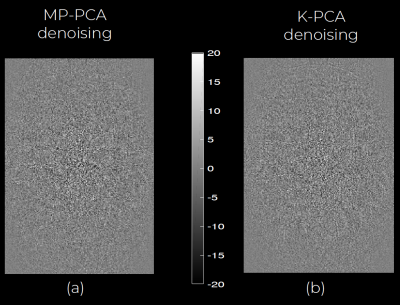
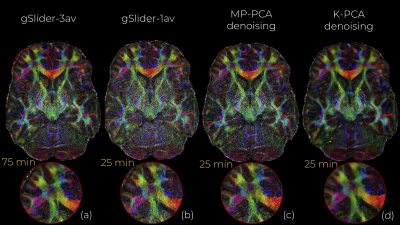
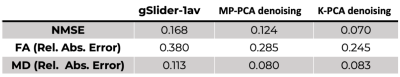
An axial slice of the denoised DWI image with MP-PCA and K-PCA is shown in Fig.1c-d, respectively, whereas the original (noisy) DWI set (gSlider-1av) is shown in Fig.1b. Clearly, K-PCA achieves substantially more noise reduction than MP-PCA. With three-times shorter acquisition time, visual quality seems superior to the gSlider-3av case (Fig.1a). This is also evident quantitatively from the NMSE results (Fig.2). Further, K-PCA has much lower NMSE in several regions compared to MP-MPCA. Residual map of K-PCA in Fig.3 does not present any structure, demonstrating that no signal information is removed. Superior noise removal can also be observed in the color-encoded FA maps that are presented in Fig 4. Fine details (see cerebellum area) remain obscured by noise in gSlider-1av and in MP-PCA. However, much better delineation of cerebellum structures, due to noise suppression, are seen with K-PCA (Fig.4d). Finally, we report (Fig.5) whole-brain errors in FA and MD. While comparable for MD, K-PCA achieves significant reduction in NMSE as well as in relative error in FA.Discussion
K-PCA can achieve a higher noise suppression than MP-PCA while preserving intricate anatomical details. An important benefit of K-PCA is that it does not require a large number of samples to perform at its best, as MP-PCA does for the MP assumption to hold. K-PCA does not need eigenvalue truncation and can work with any dMRI protocol.Conclusion
We have introduced K-PCA, a powerful and nonlinear generalization of linear PCA, that can reliably reduce noise in dMRI substantially more than state-of-the art PCA methods (MP-PCA) while preserving the underlying true signal. Showcased with 660 isotropic micrometer resolution gSlider dMRI data, we demonstrate that fine details that are obscured by noise with MP-PCA, are visible by K-PCA due to its superior noise removal performance. We believe K-PCA denoising can be an essential tool in any dMRI analysis pipeline, especially for highly noisy datasets.Acknowledgements
R01MH116173 (PIs: Setsompop, Rathi)References
1. Schölkopf, B., Smola, A., & Müller, K. R. (1997, October). Kernel principal component analysis. In International conference on artificial neural networks (pp. 583-588). Springer, Berlin, Heidelberg.
2. Mika, S., Schölkopf, B., Smola, A. J., Müller, K. R., Scholz, M., & Rätsch, G. (1999). Kernel PCA and de-noising in feature spaces. In Advances in neural information processing systems (pp. 536-542).
3. Kwok, J. Y., & Tsang, I. H. (2004). The pre-image problem in kernel methods. IEEE transactions on neural networks, 15(6), 1517-1525.
4. Veraart, J., Fieremans, E., & Novikov, D. S. (2016). Diffusion MRI noise mapping using random matrix theory. Magnetic resonance in medicine, 76(5), 1582-1593.
5. Veraart, J., Novikov, D. S., Christiaens, D., Ades-Aron, B., Sijbers, J., & Fieremans, E. (2016). Denoising of diffusion MRI using random matrix theory. NeuroImage, 142, 394-406.
6. Weinberger, K. Q., Sha, F., & Saul, L. K. (2004, July). Learning a kernel matrix for nonlinear dimensionality reduction. In Proceedings of the twenty-first international conference on Machine learning (p. 106). ACM.
7. Jorgensen, K. W., & Hansen, L. K. (2011). Model selection for Gaussian kernel PCA denoising. IEEE transactions on neural networks and learning systems, 23(1), 163-168.
8. Rathi, Y., Dambreville, S., & Tannenbaum, A. (2006, February). Statistical shape analysis using kernel PCA. In Image processing: algorithms and systems, neural networks, and machine learning (Vol. 6064, p. 60641B). International Society for Optics and Photonics.
9. Setsompop, K., Fan, Q., Stockmann, J., Bilgic, B., Huang, S., Cauley, S. F., ... & Wald, L. L. (2018). High‐resolution in vivo diffusion imaging of the human brain with generalized slice dithered enhanced resolution: Simultaneous multislice (g Slider‐SMS). Magnetic resonance in medicine, 79(1), 141-151.
Figures