0979
Prospective Motion Detection and Re-acquisition in Diffusion MRI using Phase image-based Method (PITA-MDD)1Department of Diagnostic Radiology and Nuclear Medicine, University of Maryland School of Medicine, Baltimore, MD, United States, 2Siemens Medical Solutions USA Inc, Malvern, PA, United States, 3Department of Radiology and Imaging Sciences, Indiana University School of Medicine, Indianapolis, IN, United States, 4Department of Neural and Pain Sciences and Department of Orthodontics, University of Maryland School of Dentistry, Baltimore, MD, United States, 5Deptartment of Electrical and Computer Engineering, Johns Hopkins University, Baltimore, MD, United States, 6Department of Computer Science, Johns Hopkins University, Baltimore, MD, United States
Synopsis
We have applied phase image-based motion detection (PITA-MDD) in real-time prospective motion detection and re-acquisition. During image reconstruction, PITA-MDD motion detection is performed on each slice. A diffusion-weighted volume will be re-acquired if number of motion slices exceeds the pre-set threshold. dMRI data were acquired on a volunteer using a prospective PITA-MDD sequence for the brain and the tongue. The detected motion corrupted data were consistent with subject’s motion. Denser tongue muscle fibers were visible after replacing motion volumes with re-acquired volumes. Prospective PITA-MDD motion detection and re-acquisition has improved dMRI acquisition, especially in challenging areas, such as the tongue.
INTRODUCTION
Motion artifacts cause signal loss in diffusion MRI (dMRI) images1-3 and lead to erroneous diffusion tensor estimation. Motion-free dMRI is highly desirable but is challenging to achieve especially in cases of neonatal imaging, or imaging of the tongue where involuntary motion (e.g. swallowing) is inevitable. Most dMRI analyses rely on post-processing methods to detect motion-corrupted data. For example, eddy4,5 (FSL software package) detects motion-corrupted data as outliers and either discards those data or replaces them with estimated data. Such approach causes inconsistency in useful data retained between subjects. Detecting motion during acquisition would allow for re-acquisition of corrupted views while the patient is still on the table. In this study, we extend our previous work on Phase Image Texture Analysis for Motion Detection in Diffusion MRI (PITA-MDD)6 to implement a real-time prospective motion detection and re-acquisition sequence and demonstrate the results of its performance.METHODS
Pulses sequence with online re-acquisitionThe prospective PITA-MDD sequence was designed so that phase-image based motion detection is performed online during data reconstruction, and the motion-corrupted diffusion weighted volumes are re-acquired. A prototype sequence was implemented on a Siemens platform (IDEA version VE11C). During image reconstruction, the PITA-MDD motion detection was performed per slice by calculating Haralick's homogeneity index (HHI) as a quantitative metric for homogeneity of the phase images using the following equation6,
$$$HHI=\sum_{i,j}\cfrac{p(i,j)}{1+|i-j|}$$$ (1)
where p(i,j) denotes the (i,j) element in the co-occurrence matrix of the phase image.
The re-acquisition scheme is designed as follows. Slices with HHI lower than a threshold, h, are considered as motion corrupted. A diffusion-weighted volume is subject to re-acquisition if more than N motion-corrupted slices are present. Both h and N are user-defined parameters for the sequence, which can vary depending on the object being imaged, for example, brain or tongue. Users can also define an ROI for HHI calculation for improved accuracy. HHI ROI defaults to half of imaging FOV. In tongue imaging, a user-defined box encompassing the tongue was used as HHI ROI.
Experiments
dMRI data were acquired on a volunteer using the prospective PITA-MDD sequence on a 3T scanner (MAGNETOM Prisma, Siemens Healthcare, Erlangen, Germany) for the brain and the tongue. Single Shot 2D spin-echo echo planar imaging was used with GRAPPA iPAT factor of 2, 5 B0 volumes and 63 diffusion gradient directions. FOV=240 mm, 2.5mm slice thickness, matrix size 96×96. Other imaging parameters include: Brain: 20 slices, TR=2100 ms, TE=71 ms, bandwidth=2480 Hz/Px, echo spacing 0.61 ms, b-value=1000 s/mm2, HHI threshold hbrain = 0.7, # motion slices for volume removal Nbrain = 3. Tongue: 26 slices, TR=2200 ms, TE=51 ms, bandwidth=2170 Hz/Px, echo spacing 0.55 ms, b-value=500 s/mm2, htongue = 0.5, Ntongue = 5.
The protocol was repeated twice for both brain and tongue, one with instructed motion, where the subject was instructed to move his head or swallow for three times, and the other with no instructed motion.
DTI reconstruction was performed using Diffusion Toolkit software7. Fiber tracts were extracted using TrackVis software7 with the interpolated streamline approach and an angle threshold of 35°.
RESULTS
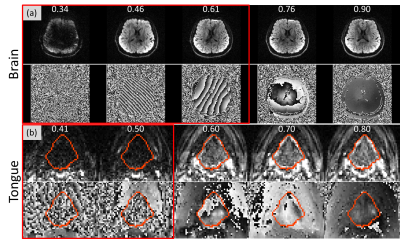
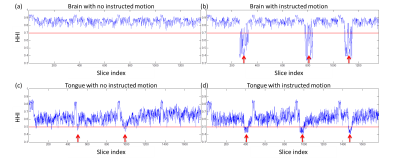
Figure 1 shows magnitude and phase images of example slices in the brain and tongue. Slices within the red boxes were motion corrupted slices detected by prospective PITA-MDD. Note that in Figure 1(a), PITA-MDD detected motion corrupted data due to head rotation (HHI = 0.46) with no apparent magnitude signal loss, which typically would be missed if one were to use magnitude-based methods. In the tongue slices (Figure 1(b)), swallowing leads to tongue signal drop-out, which is consistent with low HHI values calculated from the phase images.Figure 2 shows HHI values for all slices from the four acquisitions. Instructed head motion and swallowing are shown in Figure 2(b) and (d), represented by decreased HHI (red arrows). During the scan of the tongue with no instructed motion, the volunteer reported two instances of involuntary swallowing, which were also identified (red arrows in Figure 2(c)).
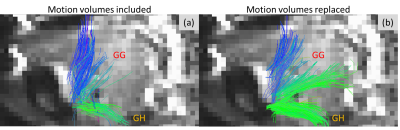
Figure 3 shows tongue muscle fibers tracked from the dMRI data of the tongue scan with instructed swallowing. Improved diffusion data quality and denser fibers were visible after replacing motion corrupted volumes with re-acquired motion-free volumes.
DISCUSSION
Implementation of the phase-based motion detection method (PITA-MDD) is feasible to be on the scanner for real-time prospective detection of motion and re-acquisition of the views that are motion corrupted in both the brain and the tongue resulting in improved dMRI data quality. Such method is especially critical in the case of tongue imaging, or other non-brain organs, where the anatomy of interest is not visible externally. We demonstrated that diffusion image quality can be drastically improved using the PITA-MDD technique in real-time. The technique does not require the use of external motion detection devices. Further optimization on the motion detection parameters are needed for HHI threshold for motion, slice threshold, and online ROI delineation for HHI calculation.CONCLUSION
We show that the phase-based motion detection technique (PITA-MDD) can be applied in real-time prospective motion detection and re-acquisition for improved dMRI acquisition in especially challenging areas, such as the tongue. Further optimization on motion detection parameters will improve the robustness of the sequence.Acknowledgements
Imaging conducted at the University of Maryland School of Medicine Center for Innovative Biomedical Resources, Translational Research in Imaging @ Maryland (CTRIM) – Baltimore, Maryland.
The work is supported by grant NIH/NIDCD R01 DC014717 and NIH/NINDS R01 NS10505503-01A1.
References
1. Anderson A, Gore J. Analysis and correction of motion artifacts in diffusion weighted imaging. Magn Res Med 1994;32:379–87.
2. Bammer R, Holdsworth S, Aksoy M, Skare S. Phase errors in diffusion-weighted imaging. In: Jones D, editor. Diffusion MRI: theory, methods, and applications. Oxford University Press; 2011. p. 218–49.
3. Trouard T, Sabharwal Y, Altbach M, Gmitro A. Analysis and comparison of motion correction technique in diffusion weighted imaging. J Magn Reson Imaging 1996;6:925–35.
4. Andersson J, Sotiropoulos S. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. NeuroImage 2016;125:1063–78.
5. Andersson J, Graham M, Zsoldos E, Sotiropoulos S. Incorporating outlier detection and replacement into a non-parametric framework for movement and distortion correction of diffusion MR images. NeuroImage 2016;141:556–72.
6. Elsaid N, Prince J, Roys S, Gullapalli R, Zhuo J. Phase Image Texture Analysis for Motion Detection in Diffusion MRI (PITA-MDD) Magn Res Imaging 2019;62:228–41.
7. Wang R, Benner T, Sorensen A, Wedeen V. Diffusion toolkit: A software package fordiffusion imaging data processing and tractography. Berlin, Germany: ISMRM;2007.
Figures