0953
Diffusion-PEPTIDE: rapid distortion-free diffusion-relaxometry imaging1A. A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, MA, United States, 2Department of Radiology, Harvard Medical School, Boston, MA, United States, 3Department of Electrical and Computer Engineering, University of Southern California, Los Angeles, CA, United States, 4Department of Biomedical Engineering, University of Southern California, Los Angeles, CA, United States, 5Harvard-MIT Health Sciences and Technology, MIT, Cambridge, MA, United States
Synopsis
Diffusion-PEPTIDE incorporates the recently developed rapid multi-shot relaxometry technique Propeller EPTI with Dynamic Encoding (PEPTIDE) into a diffusion acquisition scheme. PEPTIDE enables fast acquisition of distortion- and blurring-free images, time-resolved for different timepoints with varying T2 & T2* weighting, with self-navigation for correction of shot-to-shot phase-variation and motion. Diffusion-PEPTIDE is demonstrated here to enable distortion-free in vivo diffusion-relaxometry with large parameter space in an sensible acquisition time.
Introduction
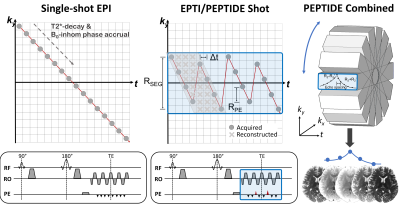
Combining diffusion with relaxometry techniques is a topic of increasing interest1,2, with the hope that it will enable investigation of sub-voxel microstructure3. However, repetition of the diffusion experiment for every change in relaxation-encoding variable (TE,TI, etc) creates impractically long acquisition durations and is a major current limitation in the field. Recent works have attempted to address this with efficient modifications to the sequence acquisition4,5, however it is still difficult to achieve a large range in both b-values and relaxation-encodings in a reasonable timeframe, particularly if combined with higher directional diffusion-encoding.A recent multi-shot EPI approach termed EPTI6 is a rapid relaxometry technique that acquires data free from the typical distortion and blurring of standard-EPI acquisitions. This has been used to acquire accurate whole-brain T2,T2*,PD and susceptibility maps from a single scan <1min. The multi-shot ky-t sampling pattern of EPTI (Figure 1) allows a B0-phase-informed GRAPPA kernel7 to reconstruct data across complete ky-t space, therefore providing a time-series of data with each image free from T2*-decay and B0-inhomogeniety-induced phase evolution effects. PEPTIDE8 is a PROPELLER9 extension to this methodology, which enables self-navigation and correction of shot-to-shot phase variation10, and has been shown to rapidly acquire quantitative information with robustness to motion of up to 30° rotation8.
This work proposes to extend the use of PEPTIDE into a diffusion acquisition scheme, utilizing its highly rapid acquisition of relaxometry data to enable joint diffusion-relaxometry in a previously infeasible acquisition duration, at high spatial-resolution and free from image distortion and blurring.
Methods
PEPTIDE sampling was incorporated into a Stejskal-Tanner diffusion sequence to achieve time-resolved distortion-free imaging, that contains multi-echo data with varying degrees of T2 and T2* weighting around the spin-echo timepoint (Figure 1). For each diffusion-direction, 12 shots of PEPTIDE blade-acquisitions are acquired to provide this distortion-free time-resolved data. The PEPTIDE specific parameters were set as previously published8 - Rseg=36, Rpe=4, ETL or Ntimepoints=72 (time-resolved images). Two diffusion-PEPTIDE protocols were tested. Protocol A: 30 directions at b=1000s/mm2 and 1 b=0 image, resolution=1.1x1.1x3.0mm3, with a TESE=108ms, TR=2.5s, ESP=1.11ms for a total scan duration of 16mins. Protocol B: 12 directions at b=500,1000,1500,2000,3000,4000,5000mm/s2 and 1 b=0 image, resolution=1.0x1.0x3.0mm3, with TESE=125ms, TR=3s, ESP=1.2ms for a total scan duration of 51mins.Three subjects were scanned. Subjects 1&2 were scanned with Protocol A and Subject 3 with Protocol B. In addition, MPRAGE and standard EPI-diffusion (R=3 in-plane acceleration, parameters otherwise matched to diffusion-PEPTIDE acquisition) data were acquired in one of the subjects, for comparing image-distortion.
Reconstruction was performed offline using MATLAB, with diffusion analysis performed in FSL11. PEPTIDE reconstruction followed the previously described pipeline8, applying phase-correction to each shot prior to combination. FSL processing was performed only to provide the diffusion modeling (“dtifit”), without any application of distortion-correction, further motion-correction, or any other typical diffusion pre-processing steps.
Results
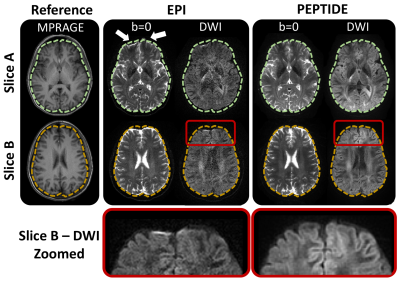
The diffusion-PEPTIDE acquisitions successfully enabled reconstruction of distortion-free time-resolved b=0 and diffusion-weighted images.Figure 2 compares the b=0 and DWI images from PEPTIDE and traditional-EPI, using the MPRAGE acquisition as a structural reference. From axial slices from different levels of the brain, typical distortion artifacts are visible for EPI whereas the PEPTIDE acquired distortion- and blurring-free images match with the structure of the reference. A zoomed-in region demonstrates that, even in regions of high EPI-distortion, PEPTIDE maintains good structural shape without the application of retrospective correction.
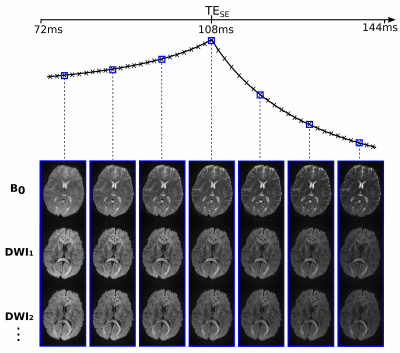
Figure 3 demonstrates the time-resolved capabilities of diffusion-PEPTIDE, showing example b=0 and DWI images for a small selection of the different reconstructed timepoints with different T2 & T2* weightings (7 out of 72).
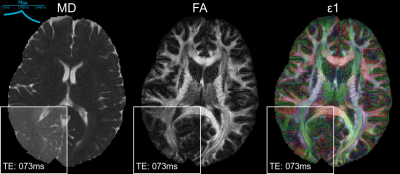
Figure 4 shows the calculated FA, MD and ε1 maps acquired using Protocol A, with a zoomed region showing the dynamic capability of these maps, cycling through the varying timepoints.
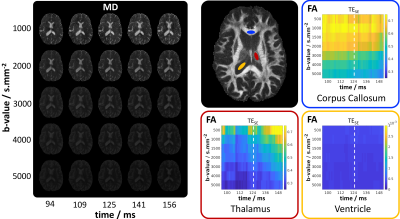
Figure 5 highlights some results for the dataset acquired with Protocol B, which enabled reconstruction of (timepoints*b-values)=(72*7)=504 diffusion datasets (each from 12 diffusion-directions). Example MD maps are shown for 5 of the b-values at 5 timepoints. On the right, plots of FA values across this dataset are demonstrated for averaged ROIs in regions associated with white-matter, gray-matter and CSF, highlighting the quantity of the potential information contained within such a dataset.
Discussion/Conclusion
PEPTIDE has been successfully incorporated into diffusion acquisitions with a reasonable acquisition time, providing distortion-free diffusion imaging capable of being resolved into a large timeseries dataset.Here we display results from a very preliminary exploration this rich diffusion-relaxometry data. FA reduces with increasing b-value as expected12, with the addition of apparent variations in FA along timepoints, differing between regions. Work is underway to utilize recent advances in diffusion-relaxation correlation modeling1,13,14 to extract interesting microstructure composition and information.
One goal of this research is to utilize the high acquisition efficiency of PEPTIDE to acquire extensive diffusion-relaxometry data to mine and better understand its limitations, for example allowing investigation into the impact of subsampling on in vivo diffusion-relaxometry spectra estimation (e.g. T2-D space1). This could potentially guide future acquisition protocols for improved focus on key parameters.
Inversion-prepared EPTI has previously been demonstrated15, which could be combined with the current approach, utilizing efficient sampling schemes such as in ZEBRA4, to provide comprehensive in vivo signal sampling (T1-T2-Diso-Ddir space), e.g. as previously performed for structural analysis of materials16,17.
Acknowledgements
This work was supported in part by NIH research grants: R01MH116173, R01EB020613, R01EB019437, U01EB025162, P41EB015896, and the shared instrumentation grants: S10RR023401, S10RR019307, S10RR019254, S10RR023043.References
1. Kim D, Doyle EK, Wisnowski JL, et al. Diffusion‐relaxation correlation spectroscopic imaging: A multidimensional approach for probing microstructure. Magn Reson Med, 78:2236-2249, 2017.
2. Veraart J, Novikov DS, Fieremans E. TE dependent Diffusion Imaging (TEdDI) distinguishes between compartmental T2 relaxation times. NeuroImage, 182:360–369, 2018.
3. Jones DK, Alexander DC, Bowtell R, et al. Microstructural imaging of the human brain with a ‘super-scanner’: 10 key advantages of ultra-strong gradients for diffusion MRI, NeuroImage, 182:8:38, 2018.
4. Hutter J, Slator PJ, Christiaens D, et al. Integrated and efficient diffusion-relaxometry using ZEBRA. Sci Rep 8:15138, 2018.
5. Ma D, Christodoulou A, Nguyen C, et al. Three-Dimensional Whole Brain Simultaneous T1, T2, and Apparent Diffusion Coefficient Mapping Using MR Multitasking. ISMRM, #0308, 2019.
6. Wang F, Dong Z, Reese TG, et al. Echo planar time‐resolved imaging (EPTI). Magn Reson Med, 81:3599– 3615, 2019.
7. Dong Z, Wang F, Reese TG, et al. Tilted‐CAIPI for highly accelerated distortion‐free EPI with point spread function (PSF) encoding. Magn Reson Med, 81: 377– 392, 2019.
8. Fair MJ, Wang F, Dong Z, et al. Propeller Echo-Planar Time-resolved Imaging with Dynamic Encoding (PEPTIDE). Magn Reson Med, in press, 2019. [DOI:10.1002/mrm.28071]
9. Pipe JG. Motion Correction with PROPELLER MRI: Application to Head Motion and Free-Breathing Cardiac Imaging. Magn Reson Med, 42:963-969, 1999.
10. Pipe JG, Farthing VG, Forbes KP. Multishot diffusion-weighted FSE using PROPELLER MRI. Magn Reson Med, 47:42-52, 2002.
11. Jenkinson M, Beckmann CF, Behrens TE, et al. FSL. NeuroImage, 62:782-90, 2012.
12. Barrio-Arranz G, de Luis-García R, Tristán-Vega A, et al. Impact of MR Acquisition Parameters on DTI Scalar Indexes: A Tractography Based Approach. PLoS One, 10(10):e0137905, 2015..
13. Kim D, Haldar JP. Faster Diffusion-Relaxation Correlation Spectroscopic Imaging (DR-CSI) using Optimized Experiment Design. ISMRM, #0176, 2017.
14. Kim D, Wisnowski JL, Haldar JP. Improved Efficiency for Microstructure Imaging using High-Dimensional MR Correlation Spectroscopic Imaging. Asilomar Conference on Signals, Systems, and Computers, p. 1264-1268, 2017.
15. Wang F, Dong Z, Reese TG, et al. 3D-EPTI for Ultra-fast Multi-contrast and Quantitative Imaging. ISMRM, #0944, 2019.
16. Topgaard D. Diffusion Tensor Distribution Imaging. NMR in Biomed, 32:4066, 2019.
17. de Almeida Martins JP, Topgaard D. Multidimensional correlation of nuclear relaxation rates and diffusion tensors for model-free investigations of heterogeneous anisotropic porous materials. Sci Rep, 8:2488, 2018.
Figures