0934
Radiomics Features of Hippocampal Regions in Conventional and Diffusion Tensor Imagings can Differentiate Temporal Lobe Epilepsy Patients
Yae Won Park1, Dongmin Choi2, Kyunghwa Han1, Sung Soo Ahn1, Hwiyoung Kim1, and Hyang Woon Lee3
1Yonsei University College of Medicine, Seoul, Republic of Korea, 2Department of Computer Science, Yonsei University, Seoul, Republic of Korea, 3Department of Neurology, Ewha Womans University College of Medicine, Seoul, Republic of Korea
1Yonsei University College of Medicine, Seoul, Republic of Korea, 2Department of Computer Science, Yonsei University, Seoul, Republic of Korea, 3Department of Neurology, Ewha Womans University College of Medicine, Seoul, Republic of Korea
Synopsis
A total of 92 subjects(66 TLE [35 right and 31 left] and 26 healthy controls) were allocated to training(n=66) and test(n=26) sets. Radiomics features (n=558) from the bilateral hippocampi were extracted from T1WI and DTI. Machine learning models were trained. Identical processes were performed to differentiate right TLE from HC and left TLE from HC. The radiomics model in test set showed better performance than hippocampal volume for identifying TLE (AUC 0.82 vs. AUC 0.62, P=0.08). Radiomics models of both subgroups showed better performance than those of hippocampal volume(AUC 0.76 vs. AUC 0.54 [P=0.12] and AUC 0.95 vs 0.68 [P=0.04]).
OBJECTIVE
To investigative whether radiomics features in bilateral hippocampi from conventional MRI and diffusion tensor imaging (DTI) can identify temporal lobe epilepsy (TLE) better than hippocampal volume.MATERIALS AND METHODS
A total of 92 subjects who had undergone MRI (66 TLE patients [35 right and 31 left TLE] and 26 healthy controls [HC]) were allocated to training (n = 66) and test (n = 26) sets. Radiomics features (n=558) from the bilateral hippocampi were extracted from MRI including T1-weighted images, mean diffusivity, and fractional anisotropy maps. After feature selection, machine learning models (random forest, extra-trees, AdaBoost, XGBoost, and LightGBM) with subsampling methods were trained. The prediction performance of the classifier was validated in the test set. Identical processes were performed to differentiate right TLE from HC (training set, n=44; test set; n=17) and left TLE from HC (training set, n=41; test set, n=16).RESULTS
The performance of the best classifier showed an area under the curve (AUC) of 0.82 in the test set to identify TLE. The radiomics model showed better performance than hippocampal volume for identifying TLE (AUC 0.62, P=0.08). The performance of the best classifiers showed an AUC of 0.76 and 0.95 in the test sets identifying right and left TLE, respectively. Radiomics models of both subgroups showed better performance than those of hippocampal volume (AUC 0.54 and AUC 0.68, with P=0.12 and P=0.04, respectively). Radiomics features were significantly correlated with neuropsychological test results.DISCUSSION
In our study, we applied radiomics analysis to identify TLE, including not only T1 images but also DTI. The performance of the multiparametric radiomics model outperformed the hippocampal volume model in the test set, showing its utility.Our multiparametric radiomics model showed a good performance (AUC 0.82) in differentiating the entire TLE from HC, indicating that radiomics has the potential for creating a generalized model that is not influenced by TLE laterality. In particular, radiomics models showed higher performance in identifying the entire TLE as well as right and left TLE, compared with hippocampal volume. Our results are in agreement with previous studies reporting that microstructural changes precede macroscopic atrophy, 1 that radiomics may reflect microstructural information different from that provided by volumetric measures.
CONCLUSION
Multiparametric radiomics classifiers may be useful imaging biomarkers for identifying temporal lobe epilepsy patients.Acknowledgements
NoneReferences
1Weston PS, Simpson IJ, Ryan NS, et al. Diffusion imaging changes in grey matter in Alzheimer’s disease: a potential marker of early neurodegeneration. Alzheimers Res Ther 2015;7(1):47.Figures
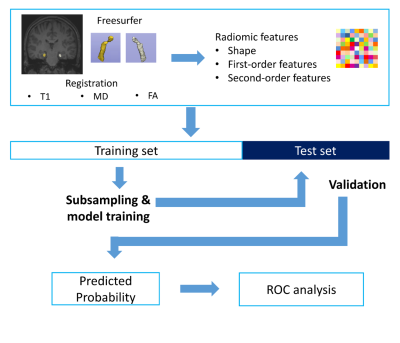
Figure 1. Workflow of image processing, radiomics feature
extraction, and machine learning
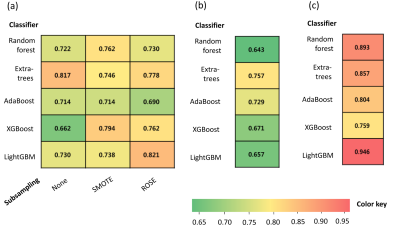
Figure
2. Heatmap of AUC values achieved from the machine learning classifiers in the test
sets for (a) differentiating TLE from HC, (b) differentiating right TLE from
HC, and (c) differentiating left TLE from HC.