0930
Deep Learning Reconstruction Method for Improved Visualization of Hippocampal Anatomical Structures1GE Healthcare, New York, NY, United States, 2Radiology, Columbia University, New York, NY, United States, 3GE Healthcare, Calgary, AB, Canada, 4Columbia University, New York, NY, United States
Synopsis
The purpose of this study was to determine if deep learning reconstruction (DLRecon) method to reduce image noise could lead to improvement in in-vivo anatomical detail of the hippocampus structures without substantial increase in scan/exam time on a clinical 3T system. Evaluation of this new reconstruction technique was performed on a group of 5 volunteers with results indicating that higher resolution scans compared to current seizure protocol was free of imaging noise and led to higher confidence in identifying hippocampal key anatomical structures and temporal lobes.
Purpose
Hippocampal high-resolution imaging is critical in the evaluation of patient with mesial temporal, localization related epilepsy, which is the most common type of epilepsy in adults. Mesial temporal sclerosis is the most common anatomic findings in adults with mesial temporal epilepsy identified on MR imaging by hyperintense T2 signal and volume loss in the hippocampus. Early findings of mesial temporal sclerosis may be subtle and are best evaluated on a 3T or higher strength magnet to obtain highest signal to noise ratio. Higher signal to noise ratio to improve spatial resolution comes at a cost of increased scan time- or increased field strength- generally causing more patient motion artifact and discomfort and overall not well tolerated.Method
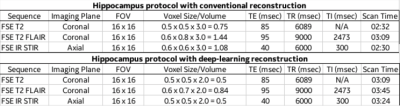
In this study, we acquired FSE coronal T2, FSE coronal T2 FLAIR and FSE axial IR sequences using current clinical voxel size compared to higher resolution/ reduced voxel size in 5 volunteers. The study was performed on, 3T 70cm bore MR scanners (Discovery MR750w, GE Healthcare, USA) using GE Signa MRI Brain Array Coil (8 channels, High Resolution). Data acquired with limited slice coverage through the anatomy of interest. Imaging parameters for deep-learning reconstruction imaging data set described in table (fig. 1). were developed by the authors to produce resolution/voxel size that would greatly improve visualization of the hippocamps compared to current seizure protocol with conventional reconstruction. The sequences collected with current clinical voxel size were reconstructed using conventional reconstruction method and the higher resolution sequences were reconstructed with the DLRecon method for removal of image noise.The Deep-Learning Reconstruction (DLRecon) method comprised a deep convolutional residual encoder network trained from a database of over 10,000 images to reconstruct images with high signal-to-noise ratio and high spatial resolution. The network offered a tunable noise reduction factor to accommodate user preference. The DLRecon network was embedded into a new reconstruction pathway that operated in parallel with the conventional reconstruction such that two sets of image series were generated from a single set of raw MR data. All deep learning reconstruction sequences used a 75% noise reduction calculation.
Results
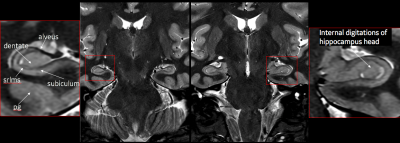
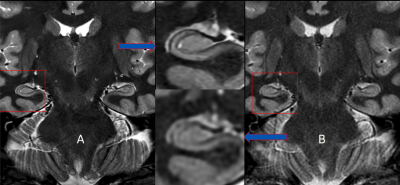
By employing the deep learning reconstruction algorithm, an increase in spatial resolution resulted in significant improvement in image quality allowing the detailed identification of hippocampal formation structures on a 3T qualitatively approaching resolution seen on a 7T magnet. This was achieved with minimal increase in scan time and without subject discomfort.Discussion
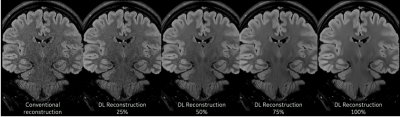
Our results from this abstract provide preliminary data on the use of DLRecon for improved visualization of the hippocampus without significant scan time increases. Increasing resolution (voxel size) in most MR sequences will improve visualization for anatomical structures when properly balanced with correct level of signal-to-noise to provide an image free, as possible, of noise. This proper balance of SNR typically comes at the expense of increase scan time, SAR and likelihood of patient motion (due to longer scan). DLRecon has shown that the use of this reconstruction method combined with protocol modifications can provide significant amount of increased SNR to produce stellar image quality with no major time penalty. This increase in SNR is key to allowing for this type of detailed imaging of the hippocampus minus the traditional methods to achieve this quality of scan (fig 2). One of the unique features of DLRecon is the flexibility for the user to dial in the specific level of noise reduction needed for a particular series (fig 3). In this study, 75% was used yielding sufficient amount of noise reduction without producing images that appear overly corrected as with filtering (low and high pass) traditionally used. This option may prove critical when dealing with data that has either high, moderate or low amount of SNR depending on the protocol.Conclusion
In this work, we demonstrated that deep learning noise removal reconstruction method can provide significant improvement to SNR and noise reduction allowing users to increase resolution for improved neuro-anatomic detail of the hippocampal formation anatomy with minimal increase in patient scan/exam time. The hippocampus is an anatomically complex structure and early hippocampal pathology remains difficult to detect in patients with temporal lobe epilepsy. This newly developed approach to noise reduction allows users to significantly shorten scan time and /or improve resolution and has many potential clinical imaging applications. For exams that require high signal to noise ratio for the highest available resolution- such as seizure protocols significant improvement in anatomic detail may be achieved without substantial scan time increase.Acknowledgements
No AcknowledgementsReferences
1. Thomas BP, Welch EB, Niederhauser BD, et al. High-resolution 7T MRI of the human hippocampus in vivo. J Magn Reson Imaging. 2008;28(5):1266–1272. doi:10.1002/jmri.21576
2. Hayman LA, Fuller GN, Cavazos JE, Pfleger MJ, Meyers CA, Jackson EF. The hippocampus: normal anatomy and pathology. AJR Am J Roentgenol 1998;171:1139–1146.
Figures