0907
A robust shimming method for in vivo abdominal mice studies based on ultrafast pulse sequences1Weizmann Institute of Science, Rehovot, Israel
Synopsis
A challenge in functional, diffusivity and spectroscopic MRI studies of mouse body regions is B0 inhomogeneity, especially at ultrahigh fields. To map and correct these DB0s, phase difference images are usually acquired using gradient echo sequences. These, however, are hard to obtain with good quality in abdominal studies due to motion artifacts. In this study we report a fully automated 3D map-based shimming method based on an ultrafast sequence that can overcome these artifacts, delivering optimal B0 homogeneity over the targeted ROIs. This technique is exemplified with mice studies at 15.2T, where its usefulness and reproducibility is demonstrated.
INTRODUCTION
Due to recent advances in genetic engineering, transgenic and knockout mice have become critical models for studying diseases. Alongside this, the availability of high and ultrahigh field magnets and cryocoils makes the sensitivity of these preclinical studies increasingly attractive. However, the small sizes involved in mouse organs and the strong susceptibility gradients arising at high fields, making it sometimes difficult to reliably obtain a sufficiently homogenous static magnetic field B0 over the regions of interest (ROIs)1,2. The optimization of B0’s homogeneity is particularly important for diffusion, functional and spectroscopic studies. Traditional iterative methods based on manually maximizing the area under an FID usually fall short of these needs, as they are time-consuming and often produce sub-optimal results. A variety of methods have therefore been proposed to provide an automated adjustment of these shims, based on a concurrent mapping of the ΔB0 inhomogeneities. One widespread such method is FASTMAP3,4, which uses six “pencils” to sample DB0 in a localized sphere. FASTMAP’s accuracy, however, like that of other methods, typically underperform when applied to motion-prone regions such as the abdomen. This study introduces a fast, fully automated 3D map-based shimming method based on spatiotemporal encoding (SPEN)5. SPEN is an ultrafast 2D MRI sequence that can overcome motion artifacts as well as certain image distortions arising from local dephasings, thanks to its possibility of performing acquisitions in a “fully-refocused” mode6 where T2* effects echo throughout the entire FID. This makes SPEN suitable for targeting challenging abdominal regions. The idea of the method here introduced is to calculate phase differences between fully-refocused and non-fully refocused SPEN 3D images, and use the ensuing multi-slice maps to calculate and correct for ΔB0 distortions. We have applied this technique to studies of the abdominal region in mouse and have evaluated its usefulness and reproducibility at 15.2T.Method
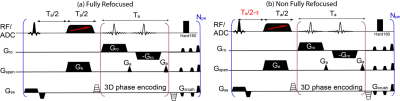
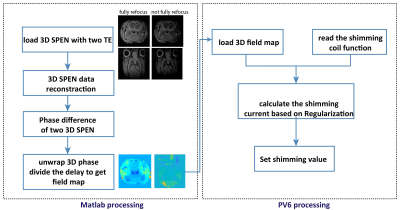
Figure 1 shows the SPEN-based pulse sequence used for B0 field mapping: Fig. 1a presents a fully refocused experiment whereby static T2* effects are refocused throughout each time point of the acquisition process –and not just at the echo center as in k-space encoding; Fig 1b introduces a ΔB0-derived the phase difference in the image by altering the echo delay from its fully-refocused, Ta/2 value. The phase difference between both SPEN experiments is thenψ=2*π*γΔB(r)*τ; on the basis of repeating this experiment over a series of phase-encoded values and processing it as described in7, volumetric maps of the DB0 can be obtained. A data acquisition protocol based on these ideas was written for a Bruker preclinical scanner running Paravision 6; the processing written for evaluating this data on the same scanner is schematized in Figure 2. The first part entails a 3D image reconstruction and 3D phase unwrapping procedure; the latter was based on phase spatial smoothness properties expected from a physical ΔB0(r), and was conducted in Matlab. The second part of the procedure involved calculating the shimming currents required to compensate the resulting ΔB0(r), and we wrote it based on a regularization algorithm provided by Bruker Paravision 6.RESULTS & DISCUSSION
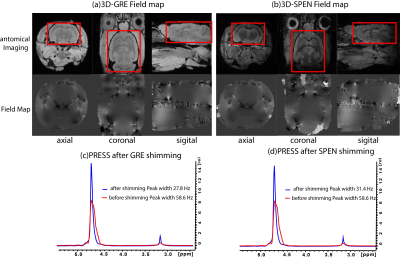
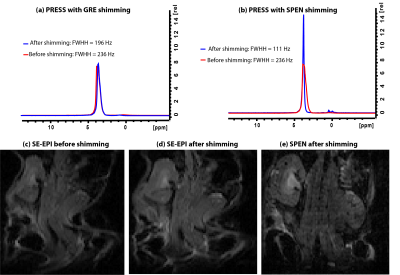
Figure 3 shows experimental results comparing our new SPEN-based field mapping method with that supplied by a conventional gradient echo approach for an in vivo mouse brain. On the upper row are the anatomical images in the different orientations, whereas in the lower row are the respective field maps. After the shimming based on SPEN pulse sequence, we can notice the width of the water peak obtained by PRESS(18*18*8 mm3 voxel) is narrowed, from 58.6 Hz to 31.4 Hz. The new method was also tested for in in vivo abdominal studies in a naïve mouse, where GRE gave poor results as a consequence of motion. Fig. 4a shows EPI images obtained before the implementation of the SPEN shimming; notice the substantial organs’ distortions. Figures 4b and 4c show the EPI and SPEN images that resulted after implementing the automated SPEN-based shimming protocol. The procedure yielded a narrowing of the abdominal water peak (PRESS, 30*30*4 mm3 voxel) from 235.5Hz to 110.8Hz. The ensuing improvement in the EPI image is evident; shown for the sake of comparison is also a single-shot 2D SPEN image collected on the same ROI at similar shimming conditions.CONCLUSION
An automatic shimming method based on ultrafast 3D SPEN was proposed and demonstrated. The advantage of the shimming method stems from its speed, its robustness to T2* effects, and its ability to overcome motion artifacts arising when using other 3D field mapping methods. The technique was found particularly valuable in mouse abdominal studies, where its usefulness and reproducibility at 15.2 T were demonstrated. Similar advantages are expected to be valid at higher preclinical fields, or in human studies at ≥7T.Acknowledgements
We are grateful to Dr. Markus Wick (Bruker BioSpin), and Mr. Koby Zibzener (Weizmann) for technical assistance. Financial support came from the NIH human placenta project (R01HD086323), the Thompson Family Foundation, the Kimmel Institute for Magnetic Resonance and the generosity of the Perlman Family Foundation.References
1. Miyasaka, N., Takahashi, K. & Hetherington, H. P. Fully automated shim mapping method for spectroscopic imaging of the mouse brain at 9.4 T. Magn. Reson. Med. 55, 198–202 (2006).
2. Hetherington, H. P., Chu, W. J., Gonen, O. & Pan, J. W. Robust fully automated shimming of the human brain for high-field 1H spectroscopic imaging. Magn. Reson. Med. 56, 26–33 (2006).
3. Gruetter, R. Automatic, localized in Vivo adjustment of all first‐and second‐order shim coils. Magnetic Resonance in Medicine 29, 804–811 (1993).
4. Gruetter, R. & Tkáč, I. Field mapping without reference scan using asymmetric echo-planar techniques. Magn. Reson. Med. 43, 319–323 (2000).
5. Tal, A. & Frydman, L. Single-scan multidimensional magnetic resonance. Prog. Nucl. Magn. Reson. Spectrosc. 57, 241–292 (2010).
6. Schmidt, R. & Frydman, L. New spatiotemporal approaches for fully refocused, multislice ultrafast 2D MRI. Magn. Reson. Med. 71, 711–722 (2014).
7. Yon, M., Bao, Q., Henriques, R., Shemesh, N. & Frydman, L. SPatiotemporal ENcoding ( SPEN ) at ultra-high fields : Applications to high resolution ( < 100 µm isotropic ) in vivo mouse brain DTI. Proc. Intl. Soc. Mag. Res. Med. 1021 17–19 (2019).
Figures