0906
A Bayesian Approach for Diffusion-Weighted Imaging to study placenta development and function in pregnancy in a large animal model1School of Biomedical Engineering & Imaging Sciences, King's College London, London, United Kingdom, 2Department of Medical Physics & Biomedical Engineering, University College London, London, United Kingdom, 3Early Origins of Adult Health Research Group, University of South Australia, Adelaide, Australia, 4Preclinical Imaging and Research Laboratories, South Australian Health and Medical Research Institute, Adelaide, Australia, 5Institute for Women's Health, University College London, London, United Kingdom, 6NIHR University College London Hospitals Biomedical Research Center, London, United Kingdom
Synopsis
Abnormalities of placental development and function result in fetal growth restriction. There is growing interest in understanding placenta structure and function throughout pregnancy to gain better understanding of placenta dysfunction. Advances in technology enables derivation of quantitative indices that reflect tissue microcapillary perfusion and tissue diffusivity from MRI. Despite recent progress, in-vivo diffusion-weighted MRI remains challenging due to long scan times, respiratory motion and low signal-to-noise ratio. Sheep provide a relevant large-scale model for invasive validation studies for MRI measurements. We aimed to improve parameter mapping using Bayesian inference. Bayesian analysis yields improved parameter maps relative to conventional least-squares fitting.
Introduction
Abnormalities of placental development and function can result in fetal growth restriction and preeclampsia1-3. A large body of evidence stresses the importance of placental insufficiency in the lifelong health of both mother and offspring4. Nevertheless, our knowledge regarding placental growth and function remains limited. Understanding the structure and function of the placenta during gestation is important to detect placental insufficiency. Advances in technology allow safe monitoring of placental growth and function in-vivo. In addition, new MR imaging techniques enable non-invasive measurement of fetal oxygen saturation5 but these have not been yet validated. Because of the invasiveness of the tests required, validation in human subjects is not possible. However, preclinical studies and testing in sheep models can define the accuracy of the MRI technique3 and its suitability for clinical use.Herein, we aimed to implement and assess Bayesian shrinkage prior (BSP) inference6,7 for placentome-specific signal modelling and compare its performance to least-squares (LSQ) fitting. Bayesian analysis yields improved parameter maps relative to conventional least-squares fitting and thus can reduce the number of individual experiments required to validate new imaging markers of placental function.
Methods
SubjectsThe study was approved by the Animal Ethics Committee of the South Australian Health and Medical Research Institute (SAHMRI) and abided by the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes developed by the National Health and Medical Research Council. Nine ewes with normal singleton pregnancy at 105-110 days gestation were included.
Image Acquisition
MRI was performed on a 3T Siemens Skyra Scanner (Erlangen, Germany). DECIDE5 imaging was performed at 11 b-values (0,10,20,30,50,70,100,150,200, 300,500,600s.mm-2) and 10 echo times (TE) (81,90,96,120,150,180,210,240,270,300 ms). All TE were acquired at b=0 to allow T2 fitting and all b-values at TE=96ms. In addition, data acquired at b=50s.mm-2 and 2000s.mm-2 for TE=(81,90,120,150,180,210,240 ms). Voxel resolution was $$$0.9 \times 0.9 \times 2.5$$$ mm.
Signal Model
Sheep placentomes comprise a 6-layer epitheliochorial structure with two inter-digitating villous capillary trees; fetal and maternal blood both remain intervascular. Based on DECIDE5, we define a sheep-specific placentome signal model:
$$S({\bf b},{\bf T_E}) = S_0 \left[ e^{-{\bf b}d^*}\left(fe^{{\bf -T_E}R_2^{f_b}}+ve^{{\bf -T_E}R_2^{mb}}\right)+(1-f-v) ve^{-{\bf b}d-{\bf T_E}R_2^{ts}}\right],$$
where $$$S$$$ is the measured MR signal and $$$S_0$$$ is the signal with b=0. The five model parameters are the fetal volume fraction $$$f$$$, diffusivity $$$d$$$, pseudo-diffusivity $$$d^*$$$, fetal blood relaxation $$$R_2^{f_b}=1/T_2^{f_b}$$$ and maternal blood volume $$$v$$$. We used literature based values for highly-saturated blood relaxation $$$R_2^{mb}=(240 ms)^{-1}$$$ and tissue relaxation $$$R_2^ts=(46ms)^{-1}$$$.
Image-Processing
A nonrigid registration algorithm8 was used to correct for motion. The sheep placentomes were manually segmented (ITK-SNAP, 2017) from the unregistered baseiline image (lowest TE, b=0).
LSQ-Fitting
The LSQ fitting is implemented in Matlab (MathWorks,Natick) using a Levenberg-Marquardt algorithm. To stabilise the voxelwise fitting the following constraints were chosen: $$$0<f<1$$$ (no units), $$$0<d<1 $$$ $$$(mm^2s^-1)$$$, $$$0<d*<1$$$ $$$(mm^2s^-1)$$$, $$$0<$$$T2 fetal blood$$$< 240$$$ (ms), $$$0<v<1$$$ (no units).
BSP Inference
Our method6uses a hierarchical prior in which spatial correlation is introduced by modelling regions as containing voxels with similar values. This correlation is incorporated in the parameter inference leading to large-scale spatial smoothness. We used a Monte Carlo Markov Chain approach to perform inference using Gibbs sampling. This is initialised with the voxel-wise LSQ estimates.
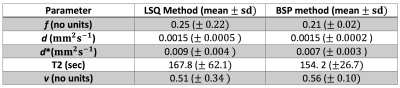
The effect of BSP method was evaluated by visual comparison of parameter maps calculated with BSP and LSQ methods. Mean value and standard deviation were calculated over the 9 subjects.
Results
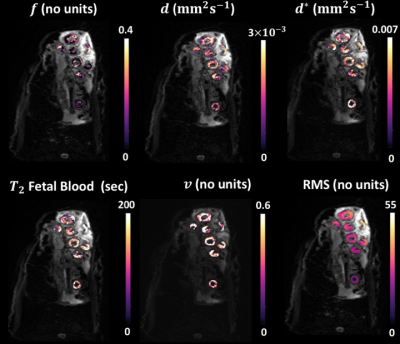
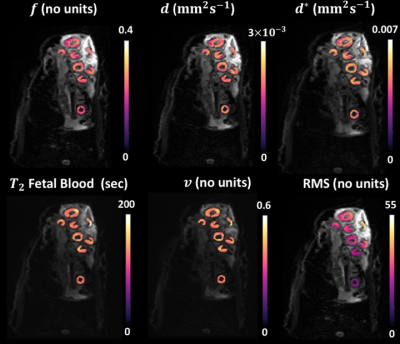
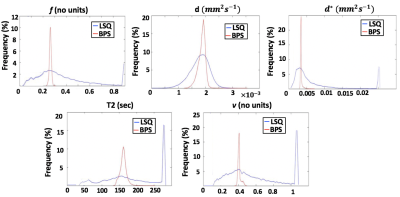
Fig.1 and Fig.2 show examples of the parameter maps obtained with the LSQ and BSP approaches respectively. All LSQ parameter maps appear noisy and artefact-prone, where BSP fitting notably improved all the parameter maps. Fig.3 shows corresponding histograms of the 5 parameters calculated with LSQ and BSP method. The proportions of the LSQ estimated in the edge-most bins are 4% for $$$f$$$, 7.1% for $$$d^{*}$$$, 17.3% for T2 and 19.5% for $$$v$$$. ROI mean estimated values derived using BSP approach are comparable to LSQ estimates but the precision of the parameters has been improved.Discussion
To minimise variability related to noise we evaluated our data averaged over all ROIs drawn around placentomes. Even then, applying LSQ fitting resulted in poor measurement of the parameters. The improvement in BSP derived parameter maps was typical for all the in-vivo sheep placentomes. The shrinkage effect of the prior distribution in BSP approach is clearly seen in Fig.3 where the outlying LSQ estimates have been brought into the bulk of the distribution. In contrast to LSQ method which allows data processing in a pixel-wise fashion, BSP approach requires pre-segmentation of the data which makes automation in image-processing workflow more challenging. The BSP approach is applicable to many forms of image contrast where model fitting is necessary. Further work is needed to evaluate the effect of the BSP method on the diagnostic quality of the estimated parameter maps with histological data.Conclusion
BSP method yields improved parameter maps relative to conventional LSQ fitting in the sheep placentomes. Further in-vivo studies are necessary to assess the performance of the method at a wider range of gestational ages. This information will help demonstrate the longitudinal trend between the MR parameters during pregnancy that may aid in the assessment of novel treatments for placental dysfunction.Acknowledgements
This research was supported by the Wellcome Trust (210182/Z/18/Z), Wellcome Trust/EPSRC (NS/A000027/1) and the Radiological Research Trust.References
- Scifre CM and Nelson DM. Intrauterine growth restriction, human placental development and trophoblast cell death. J Physiol. 2019; 587(Pt 14): 3453 – 3458.
- Swanson AM, David A. Animal models of fetal growth restriction: Considerations for translational medicine. Placenta. 2015; 36(6):623-630.
- Morrison, JL . Sheep models of intrauterine growth restriction: fetal adaptations and consequences. Clin Exp Pharmacol Physiol. 2008; 35(7):730–43.
- Spencer RN, Carr DJ and David AL. Treatment of poor placentation and the prevention of associated adverse outcomes – what does the future hold?. Prenat Diagn. 2014; 34: 677–684.
- Melbourne A, Aughwane R, Sokolska M, et al. Separating fetal and maternal placenta circulations using multiparametric MRI. Magn Reson Med. 2018; 00:1–12.
- Flouri D, Owen D, Aughwane R, et al. Improved Placental Parameter Estimation Using Data-Driven Bayesian Modelling. Med Image Comput Comput Assist Interv. 2019; 11766:609–616.
- Orton MR, Collins DJ, Koh DM et al. Improved Intravoxel Incoherent Motion Analysis of Diffusion Weighted Imaging by Data Driven Bayesian Modeling. Magn Reson Med. 2014; 71: 411–420.
- Melbourne A, Atkinson D, White MJ, et al. Registration of dynamic contrast‐enhanced MRI using a progressive principal component registration (PPCR). Phys Med Biol. 2007; 52:5147–5156.
Figures