0894
Deep Learning-based Single-shot T2* Mapping Using Multiple Overlapping-Echo Detachment Acquisition1Department of Electronics Science, Xiamen University, Xiamen, Fujian, China
Synopsis
Quantitative MRI is of great value to both clinical diagnosis and scientific research. In this study, a novel T2* mapping method, gradient-echo multiple overlapping-echo detachment acquisition (GRE-MOLED) sequence with deep learning-based reconstruction algorithm was proposed. The method is capable of acquiring reliable T2*, M0 and B0 maps simultaneously in a single shot and is robust to B0-inhomogeneities. As a rapid T2* quantitative tool, GRE-MOLED reduces the scan time of T2* mapping to less than 75 ms per slice and has great potential in clinical real-time applications.
Introduction
Quantification of T2* is of major clinical and research interest, with applications including blood oxygenation level-dependent (BOLD) fMRI1, dynamic perfusion MRI2 and detection of iron deposition to assessment of patients suffering from Parkinson’s3 and Alzheimer’s4 diseases. Long acquisition time makes the T2* mapping vulnerable to the effect of motion and is not suitable for dynamic quantitative imaging. In this work, we develop a new approach, gradient-echo multiple overlapping-echo detachment acquisition (GRE-MOLED) sequence, to achieve fast quantification of T2*, M0 and B0 simultaneously in a single shot. Meanwhile, with a deep learning-based reconstruction algorithm, the approach is capable of presenting images immune to distortion.Methods
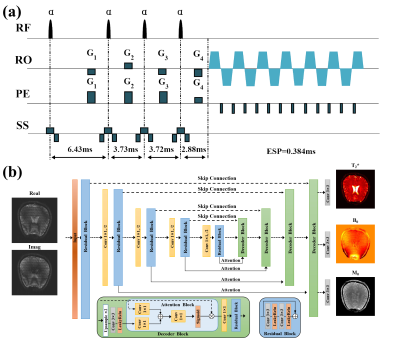
The proposed single-shot GRE-MOLED sequence is shown in Fig. 1a. Compared with the original MOLED sequence,5,6 the refocusing pulse is removed but four excitation pulses with flip angle α are retained to achieve four echoes with different T2*-weighting. For network training, 5000 simulated training samples with size of 256×256 were randomly generated by SPROM software. The pulse sequence and parameters used in simulation were the same as those used in experiments.All data were obtained on a 3T MRI system. The reference T2* maps were fitted from a series of images acquired with the gradient echo based sequence with different TE values. A GRAPPA acceleration rate of 2 was used to accelerate the acquisition. The pulse flip angle α was set to 15°.
After acquisition, both the raw data from numerical simulation and in vivo human brain are reconstructed using an encoder-decoder network, residual attention UNet (ResAttUNet), which consists of attention block and residual learning block (Fig. 1b). The ResAttUNet is used to learn the non-linear mapping between patches of raw data and the corresponding T2*, M0 and B0 maps. The input has been decomposed into real and imaginary components to enable complex-valued processing for reconstruction. During training, a multi-task learning strategy is introduced to reconstruct T2*, M0 and B0 maps simultaneously by performing a weighted linear sum of the losses for each individual task as follows:
$$ L_{total}=\sum_iw_{i}L_{i} $$
where w is weight of the i-th task and L represents pixel-wise mean squared error (MSE). We set wT2*:wM0:wB0=1:0.5:0.2. At the testing stage, experimental data were fed into the trained ResAttUNet to reconstruct the parameter maps.
Results
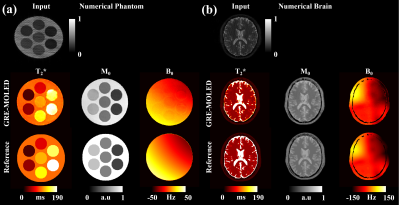
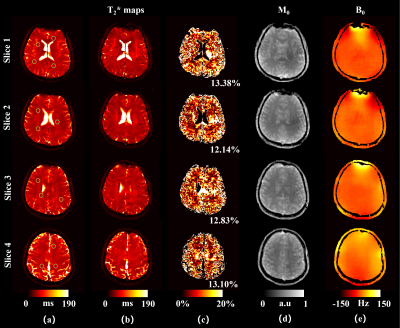
Figure 2 illustrates the results of a numerical phantom and numerical brain. The T2* and M0, B0 maps of GRE-MOLED are very close to the corresponding references, and the image distortion is rectified by our network model. Compared with the input images acquired, the reconstructed parameter maps shows strong robustness to B0 inhomogeneity (-50 Hz – 50 Hz for phantom and -150 Hz – 150 Hz for brain).Figure 3 illustrates the results of representative slices from a human brain in vivo experiments. The scan time is 75 ms per slice for GRE-MOLED sequence, while is about 6 s per slice for GRE sequence with four echoes. The Pearson’s correlation coefficients of the T2* values of all pixels in the ROIs (green circles in Fig. 3a) between the references and the reconstructed results are larger than 0.989. The mean relative error of all these 21 slices is about 12.74%. Meanwhile, the results of M0 and B0 also show high quality in Fig. 3d and 3e.
Discussion
The proposed GRE-MOLED sequence and reconstruction method show excellent performance in both simulation and in vivo brain experiments. Compared with traditional GRE sequence, GRE-MOLED can yield higher temporal resolution which makes it applicable for dynamic quantitative imaging. Meanwhile, the ResAttUNet network shows a strong nonlinear fitting and multi-task learning capabilities. Due to the large receptive field of the encoder-decoder architecture, the image distortion can be rectified. In addition, owing to a large T2* range (20 ms – 300 ms), spatial attention mechanism is introduced in the decoder block to prevent the network from continuously optimizing the weights that are beneficial to large T2* values in the training process.Conclusion
In this study, we proposed a novel single-shot T2* mapping method, GRE-MOLED based on MOLED sequence. It can be used to simultaneously quantify T2*, M0, and B0. It has great potential in clinical applications.Acknowledgements
This work was supported by the National Natural Science Foundation of China under grant numbers 11775184 and 81671674.References
1. Jochimsen TH, Möller HE. Increasing specificity in functional magnetic resonance imaging by estimation of vessel size based on changes in blood oxygenation. NeuroImage. 2008;40:228–236.
2. Johnson G, Wetzel SG, Cha S, Babb J, Tofts PS. Measuring blood volume and vascular transfer constant from dynamic, t2*‐weighted contrast‐enhanced MRI. Magn. Reson. Med. 2004; 51( 5): 961–968.
3. Vymazal J, Righini A, Brooks RA, Canesi M, Mariani C, Leonardi M, et al. T1 and T2 in the brain of healthy subjects, patients with Parkinson disease, and patients with multiple system atrophy: relation to iron content. Radiology 1999;211:489–495.
4. Ordidge RJ, Gorell JM, Deniau JC, Knight RA, Helpern JA. Assessment of relative brain iron concentrations using T2-weighted and T2*- weighted MRI at 3 Tesla. Magn Reson Med 1994;32:335–341.
5. Cai CB, Zeng YQ, Zhuang YC, et al. Single-shot T2 mapping through overlapping-echo detachment (OLED) planar imaging. IEEE Trans. Biomed. Engineering, 2017; 64: 2450-2461.
6. Zhang J, Wu J, Chen SJ, et al. Robust single-shot T2 mapping via multiple overlapping-echo acquisition and deep neural network. IEEE Trans. Med. Imaging, 2019; 38: 1801-1811.
Figures