0889
Accelerated T2 Mapping by Integrating Two-Stage Learning with Sparse Modeling
Ziyu Meng1,2, Yudu Li2,3, Rong Guo2,3, Yibo Zhao2,3, Tianyao Wang4, Fanyang Yu2,5, Brad Sutton2,5, Yao Li1, and Zhi-Pei Liang2,3
1Institute for Medical Imaging Technology, School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China, 2Beckman Institute of Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 3Department of Electrical and Computer Engineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 4Department of Radiology, The Fifth People's Hospital of Shanghai, Fudan University, Shanghai, China, 5Department of Bioengineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States
1Institute for Medical Imaging Technology, School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China, 2Beckman Institute of Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 3Department of Electrical and Computer Engineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 4Department of Radiology, The Fifth People's Hospital of Shanghai, Fudan University, Shanghai, China, 5Department of Bioengineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States
Synopsis
We propose a new method to learn the multi-TE image priors for accelerated T2 mapping. The proposed method has the following key features: a) fully leveraging the Human Connectome Project (HCP) database to learn T2-weighted image priors for a single TE, b) transferring the learned single-TE T2-weighted image priors to multi-TE via deep histogram mapping, c) reducing the learning complexity using a tissue-based training strategy, and d) recovering subject-dependent novel features using sparse modeling. The proposed method has been validated using experimental data, producing very encouraging results.
Introduction
Quantitative T2 mapping requires acquisition of multi-TE T2-weighted (T2W) data, often leading to a long scan time. To accelerate T2 mapping, many methods have been developed, including the more recent deep-learning (DL) based methods1-6. DL-based methods attempt to learn a high-dimensional functional, which requires large amounts of training data for stable solutions7. However, sufficient training data for different TEs are not publicly available, thus existing deep networks can only generate image priors for a single TE. To address this issue, we propose a new method to learn and incorporate multi-TE image priors for accelerated T2 mapping, characterized by: a) fully leveraging the Human Connectome Project (HCP) database to learn T2-weighted image priors for a single TE, b) transferring the image priors learned from the HCP data to multiple TEs via deep histogram mapping, c) reducing the learning complexity using a tissue-based training strategy, and d) recovering subject-dependent novel features using sparse modeling. The proposed method has been validated using experimental data, generating very encouraging results.Methods
Learning multi-TE image priors using a two-stage deep learningWe proposed a novel two-stage DL strategy to learn multi-TE image priors, whose key elements are summarized in Fig. 1. At stage 1, we leveraged the large number of training data in the HCP database and learned the single-TE (effective TE = 565 ms) T2W image priors using a T1W-T2W image translation network. At stage 2, we transferred the learned single-TE priors to multi-TE (TE = 12, 116, 303 ms) images using separate networks for nonlinear histogram/distribution remapping. In addition, we applied a novel tissue-based training strategy for all the networks involved (i.e., trained the deep networks tissue by tissue) to reduce the dimensionality and complexity of the functions to be learned.
In our preliminary implementation, we used one of the state-of-the-art image translation network architectures, pix2pixGAN8, in both learning stages. For T1W-T2W image translation, we used 900, 100 and 100 datasets in HCP database for training, validation and testing, respectively. For the single-TE to multi-TE transferring, we acquired a new set of training data from 23 subjects. All these training data were collected on a 3T scanner with the following imaging protocols: FSE (FOV: 240x240x72 mm3, matrix size: 256x256x24, TE: 12,116, 303 ms, TR: 1100 ms) for multi-TE T2W images and MRPAGE (FOV: 240x240x192 mm3, matrix size: 256x256x192, TE/TI/TR: 2.29/900/1900 ms) for T1W images.
Recovering the subject-dependent novel features by sparse modeling
In practice, directly using the DL-based image priors as the final solutions can introduce significant biases, especially in the presence of novel features such as lesions. In this work, we used a novel sparse model to absorb the DL-based image priors and capture the subject-dependent novel features. Particularly, for each TE, we solved the following constrained optimization problem (ignoring TE for simplicity):
$$\{\hat{c}_m\},\hat{\rho}_{\mathrm{n}}=\arg\min_{\{c_m\},\rho_{\mathrm{n}}}\left\|d-\Omega\mathcal{F}\left(\rho_{\mathrm{ML}}(\boldsymbol{x})\left(\sum_{m=-M/2+1}^{M/2}c_me^{i2\pi m\Delta\boldsymbol{k}\boldsymbol{x}}\right)+\rho_{\mathrm{n}}(\boldsymbol{x})\right)\right\|_2^2+\lambda\|W\rho_{\mathrm{n}}(\boldsymbol{x})\|_1$$
where $$$d$$$ is the measured data, $$$\Omega$$$ the sampling operator and $$$\mathcal{F}$$$ the Fourier operator. The learned image prior $$$\rho_{\mathrm{ML}}(\boldsymbol{x})$$$ is incorporated into the solution using the generalized series (GS) model9 and $$$\rho_{\mathrm{n}}(\boldsymbol{x})$$$ is a sparse term under some sparsifying transform $$$W$$$. The purpose of the GS model is to compensate for the smooth discrepancies between the desired image function and $$$\rho_{\mathrm{ML}}(\boldsymbol{x})$$$, while $$$\rho_{\mathrm{n}}(\boldsymbol{x})$$$ is to capture localized novel features (e.g., lesions). After reconstruction for all TEs, T2 maps were calculated using the standard single-component exponential fitting procedure2.
Results
Figure 2 shows some representative results of the effect of training data size on the learning outcome for T2 mapping. As shown in Fig. 2(a), DL produced satisfactory predictions if trained with adequate training samples, but performed significantly worse with limited training data due to the overfitting problem. A common strategy to alleviate this problem is to use data augmentation by combining data from all the slices across the brain but at the cost of severe blurring artifacts, as shown in Fig. 2(b).To evaluate the proposed strategy for learning multi-TE image priors, we compared it with the existing training strategies4 that only used the acquired limited multi-TE training data without leveraging the HCP database. As shown in Fig 3, the proposed method significantly outperformed the traditional methods with reduced image blurring and artifacts.
The performance of the proposed image reconstruction method incorporating the learned multi-TE priors was validated in accelerated T2 mapping. To this end, we retrospectively under-sampled the acquired multi-TE data, using a random sparse sampling scheme with 12.5% sparsity (Fig. 4(a)). As shown in Fig. 4(b), the learned image priors helped considerably reduce the spatial aliasing artifacts, compared to the conventional compressive-sensing-based reconstruction. In addition, the proposed method successfully recovered the subject-dependent novel features, as shown in Fig. 5(a). The proposed method produced impressive T2 maps shown in Fig. 5(b).
Conclusions
This paper presents a new method for accelerating T2 mapping by effectively integrating a two-stage DL approach with a sparse model. The proposed method fully takes advantage of the HCP database to obtain multi-TE T2W image priors and capture local novel features by sparse modeling. The performance of the proposed method has been evaluated using experimental data, producing encouraging results. With further development, the method may prove useful for many applications involving T2 mapping.Acknowledgements
This work was supported, in part, by NIH-R21-EB023413 and NIH-U01-EB026978.References
- Doneva M, Börnert P, Eggers H, et al. Compressed sensing reconstruction for magnetic resonance parameter mapping. Magn Reson Med. 2010;64(4):1114-1120.
- Zhao B, Lu W, Hitchens TK, et al. Accelerated MR parameter mapping with low‐rank and sparsity constraints. Magn Reson Med. 2015;74(2):489-498.
- Lee D, Jin KH, Kim EY, et al. Acceleration of MR parameter mapping using annihilating filter‐based low rank hankel matrix (ALOHA). Magn Reson Med. 2016;76(6):1848-1864.
- Xiang L, Chen Y, Chang W, et al. Deep Leaning Based Multi-Modal Fusion for Fast MR Reconstruction. IEEE Trans Biomed Eng. 2019;66(7):2105-2114.
- Cai C, Wang C, Zeng Y, et al. Single‐shot T2 mapping using overlapping‐echo detachment planar imaging and a deep convolutional neural network. Magn Reson Med. 2018;80(5):2202-2214.
- Liu F, Feng L, Kijowski R. MANTIS: Model‐Augmented Neural neTwork with Incoherent k‐space Sampling for efficient MR parameter mapping. Magn Reson Med. 2018;82(1):174-188.
- Antun V, Renna F, Poon C, et al. On instabilities of deep learning in image reconstruction-Does AI come at a cost? arXiv preprint arXiv:1902.05300. 2019.
- Isola P, Zhu JY, Zhou T, et al. Image-to-image translation with conditional adversarial networks. In Proc. Conf. Comput. Vis., 2017.
- Liang ZP, Lauterbur PC. A generalized series approach to MR spectroscopic imaging. IEEE Trans Med Imaging. 1991;10(2):132-137.
Figures

Figure
1: Illustration of the proposed two-stage deep learning strategy. At stage 1,
we leveraged the HCP database to learn T2W image priors for a single TE (TE 565
ms) using a T1W-T2W image translation network. At stage 2, we transferred the
learned T2W image priors to multiple TEs (TE = 12, 116, 303ms) using deep histogram
remapping. The training tasks at each stage were accomplished using tissue-based
learning.

Figure
2: Effect of training data size on traditional
DL-based T1W-T2W image translation. (a) In this experiment, the T1W-T2W translation
network was trained using different numbers of training data. As can be seen,
DL produced satisfactory predictions if trained with adequate training samples,
but performed poorly with limited training data due to overfitting. (b) Illustration
of the limitation in data augmentation using all slices across the brain for
training. As can be seen, the network thus trained produced significant
blurring artifacts.
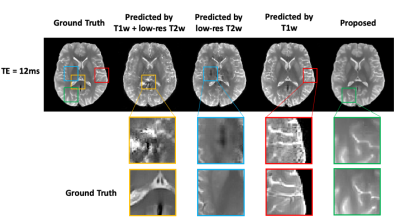
Figure
3. Comparison of the proposed method with traditional DL-based methods. Note
that the predictions from the traditional method suffered from overfitting and
showed significant blurring and artifacts due to limited training data, which
is consistent with the observations from Fig. 2. These artifacts were
significantly reduced by the proposed method.

Figure
4. Illustration of accelerated T2 mapping using the learned multi-TE T2W image priors. We retrospectively under-sampled the acquired multi-TE data,
using a random sparse sampling scheme with 12.5% sparsity, as shown in Fig. 4(a).
Compared with the conventional compressive-sensing-based method, the proposed
method produced much better T2 maps with relatively high acceleration factor
(sparsity = 12.5%).
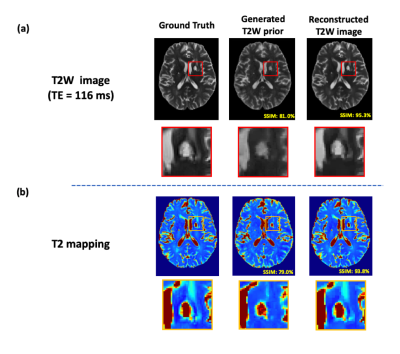
Figure
5. Illustration of novel feature reconstruction by integrating learned T2W
priors with sparse modeling. Using an example data from a patient with a lacunar infarction, Fig. 5(a) indicates that the proposed sparse modeling can successfully
recover the subject-dependent novel features. Fig. 5(b) demonstrates that
direct use of DL-based image priors as the final solutions will introduce
significant biases, especially in the presence of novel features such as
lesions.