0859
Lesion-Robust White-Matter Bundle Identification through Diffusion Driven Label Fusion1Neuropsycology, Max Planck Institute for Human Cognitive and Brain Sciences, Leipzig, Germany, 2Parietal, Inria Saclay - Ile de France, PAris, France, 3Athena EPI, Universite Cote d’Azur, Inria, Sophia Antipolis, France, 4Psychiatry Neuroimaging Laboratory, Brigham and Womens Hospital, Harvard Medical School, Boston, MA, United States, 5Max Planck Institute for Human Cognitive and Brain Sciences, Leipzig, Germany
Synopsis
White-matter pathologies disrupt the white-matter organization, that manifests as deficits in brain function. When treating such pathologies, it is of great importance to infer which pathways are affected. However, the white-matter lesions hamper the use of tractography to track fiber bundles. In this work, leveraging diffusion imaging, we propose a novel diffusion-driven technique to improve the localization of brain pathways. Aggregating information from few healthy subjects, our technique is able to localize both the affected pathways and the lesion interrupting when tracking is not possible.
Introduction
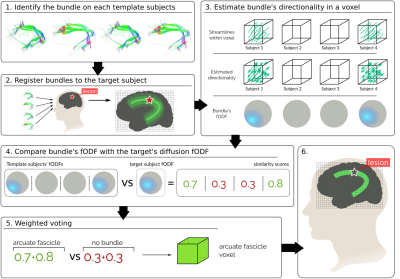
Pathologies such as infiltrative tumors disrupt the structure of white matter in the brain, resulting in cognitive deficits. Inferring which pathways are affected by the lesion is key for both pre and post-treatment planning. In the presence of lesions, tractography algorithms fail to characterize white matter pathways[1]. We solve this by harnessing aggregated healthy subject information. Given a set of labeled major bundles in a group of healthy subjects, we non-linearly register them to our patient’s brain and combine them using a label fusion algorithm. A major advantage of label fusion techniques is their high accuracy even when inferring from few subjects[2]. Current label fusion techniques rely on tissue contrast, not taking into account the fibrous structure of white matter[2]. Leveraging that brain structures constrain the diffusion of water particles differently, we propose a novel diffusion-driven technique to improve the localization of brain pathways (Fig. 1). We show its feasibility and advantages in both synthetic and human data.Methods
Let $$$\{L_s\},s\in S$$$ be a set of streamlines in a common space representing white-matter pathways of template subjects $$$S$$$. Let $$$\{l_i\}$$$ be the set of labels representing those pathways in the brain. We propose a weighted voting technique to label the voxel $$$x$$$ of a target subject:\begin{equation*}\label{eq:mvoting_weighted}L^*(x)=argmax_{l\in labels}\sum_{s\in S}p(L(x)=l|L_s(x))p(D(x)|D_{sl}(x))\end{equation*}
\begin{equation*}\begin{aligned}p(L(x)=l|L_s(x))=\begin{cases}1,&\text{if }L_s(x)=l\\0,&\text{otherwise}\end{cases}\end{aligned}\end{equation*}
\begin{equation*}\begin{aligned}p(D(x)|D_{sl}(x))=\begin{cases}\langle F(x),F_{sl}(x)\rangle,&\text{if }L_s(x)=l,\text{and }l\neq0\\\langle F(x),U\rangle,&\text{if }L_s(x)=0\\0,&\text{otherwise.}\end{cases}\end{aligned}\end{equation*}
The term $$$p(L(x)=l|L_s(x))$$$ represents the "vote" of each template[3], being 1 if the bundle $$$l$$$ is present in the template's voxel $$$x$$$ and 0 if not. The term $$$p(D(x)|D_{sl}(x))$$$ weighs the vote based on how much the voted bundle resembles the target's diffusion data. It express the probability of seeing the target's diffusion, $$$D(x)$$$, given that the template's bundle $$$l$$$ is present in the voxel, $$$D_{sl}(x)$$$.
We characterize $$$D(x)$$$ by fitting a Constrained Spherical Deconvolution (CSD) model[4] to the target's diffusion data and estimating a fiber orientation density function (fODF). The fODF $$$F_x(\theta, \phi)$$$ represents the fraction of fibers within the voxel $$$x$$$ aligned along the spherical coordinate $$$(\theta, \phi)$$$. Then, we characterize the within-voxel directionality of the template's bundle by looking at the entry and exit points (Fig. 1.3) of its streamlines. We estimate a fODF from these directions by means of CSD as with the diffusion data. If the template has no streamlines in the voxel, a uniform fODF, $$$U$$$, is used. Finally, we define $$$p(D(x)|D_{sl}(x))$$$ as the inner product between the diffusion-based fODF, $$$F(x)$$$, and the bundle fODF, $$$F_{sl}(x)$$$, with both fODFs normalized such that $$$\langle F(x),F(x)\rangle=1$$$.
Experiments and Results
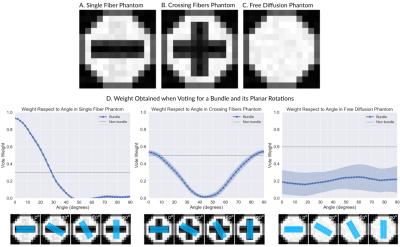
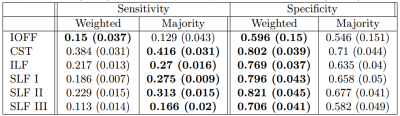
Our technique performs weighted voting, were votes for bundles aligned with the target's diffusion signal will get higher votes. To assess that our technique weighs the votes correctly we created 3 synthetic diffusion datasets using Phantomas[5]. The first phantom possesses only one bundle, the second possesses two crossing bundles and the third no bundles (Fig. 2A-C). We generated 30 DWIs per phantom using an SNR=20, a resolution of 1mm per voxel, 4 bvalues (0, 1000, 2000, and 3000), and 200 directions. We used the DWIs to computed the weight a vote for a bundle and its planer rotations would obtain (Fig. 2D). Figure 2D shows that in the first phantom the weight starts to decrease as the angle increments and the directionality of the bundle moves away from that of the diffusion. In the second phantom, the weight is higher when the bundle aligns with one of the crossing fibers (at 0 or 90 degrees). In the third phantom, the bundle's vote weight is always lower than the one for the "non bundle" label.To assess the benefits of adding a weight to the votes, we tested our technique in a more realistic scenario. We randomly selected 10 subjects from the Human Connectome Project, computed whole-brain tractography in them and extracted 6 bundles using the white-matter query language (WMQL)[6]. For each bundle we performed a leave-one-out cross-validation experiment. At each step, we inferred the bundle of one subject from the registered bundles of the others using our technique with and without weights. Using as ``ground truth'' the target's bundle obtained with WMQL, we quantified the sensitivity and specificity of both techniques (Table 1). In all bundles adding the weight achieves a lower sensitivity but a greater specificity. Therefore, we label fewer voxels, but those which are labeled can be trusted more.
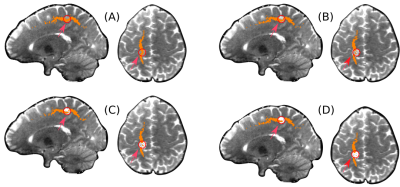
To compare how the labeling behaves in the presence of lesions, we simulated a disruptive lesion in the Superior Longitudinal Fascicle (SLF) bundle (Fig. 3). We did so by selecting a spherical region of 4mm radius where the SLF passes and mixed the diffusion signal with isotropic diffusion. Figure 3 shows that the more isotropic the signal, the fewer voxels are labeled within the lesion, allowing to identify the lesion within the bundle.
Conclusion
We presented a label fusion technique to localize fiber bundles when whole-bundle tracking is hampered by non-deforming lesions. Through 3 experiments we showed that our technique: weights votes correctly; is more conservative than the traditional majority voting, a desirable feature in clinical applications; and contrary to normal voting, it does not label voxels within a lesion, allowing to identify both the affected bundle and the lesion interrupting it.Acknowledgements
This work was partially financed by the grant R01HD090641, the ANR NeuroRef and the ERC-StG NeuroLang. This work has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (ERC Advanced Grant agreement No 694665 : CoBCoM - Computational Brain Connectivity Mapping).References
[1] B. J. Jellison, A. S. Field, J. Medow, M. Lazar, M. S. Salamat, and A. L. Alexander, “Diffusion Tensor Imaging of Cerebral White Matter: A Pictorial Review of Physics, Fiber Tract Anatomy, and Tumor Imaging Patterns,” American Journal of Neuroradiology, 2004.
[2] A. J. Asman and B. A. Landman, “Non-local statistical label fusion for multi-atlas segmentation,” Med. Image Anal., vol. 17, no. 2, pp. 194–208, 2013
[3] L. Xu, A. Krzyżak, and C. Y. Suen, “Methods of Combining Multiple Classifiers and Their Applications to Handwriting Recognition,” IEEE Trans. Syst. Man Cybern., vol. 22, no. 3, pp. 418–435, 1992
[4] J.-D. Tournier, F. Calamante, D. G. Gadian, and A. Connelly, “Direct estimation of the fiber orientation density function from diffusion-weighted MRI data using spherical deconvolution,” Neuroimage, vol. 23, pp. 1176–1185, Nov 2004
[5] E. Caruyer, A. Daducci, M. Descoteaux, J.-C. Houde, J.-P. Thiran, and R. Verma, “Phantomas: a flexible software library to simulate diffusion MR phantoms,” Ismrm, vol. 17, p. 20013, 2014.
[6] D. Wassermann, N. Makris, Y. Rathi, M. Shenton, R. Kikinis, M. Kubicki, and C.-f. Westin, “The white matter query language: a novel approach for describing human white matter anatomy,” Brain Struct. Funct., vol. 221, pp. 4705–4721, dec 2016.
Figures