0851
Tract Dictionary Learning for Fast and Robust Recognition of Fiber Bundles1Department of Radiology and BRIC, University of North Carolina, Chapel Hill, Chapel Hill, NC, United States
Synopsis
Fiber bundle parcellation is key to bundle-specific analysis of white matter pathways.In this abstract, we propose an efficient framework for parcellation of white matter tractograms using discriminative dictionary learning. The key to our framework is to learn a compact dictionary for each fiber bundle so that the streamlines within the bundle can be succinctly represented. Experiments on a bundle-labeled HCP dataset and an infant dataset highlight the ability of our framework in grouping streamlines into anatomically plausible bundles.
Purpose
Diffusion MRI (dMRI)1 allows the analysis of white matter (WM) axonal trajectories via streamlines (i.e., tractograms) generated via a process called tractography. Automatic grouping of several millions of tractograms into anatomically meaningful bundles is an important but challenging 2,3 since tractograms within a bundle can have different shapes, lengths, and endpoints. Utilizing standard geometric distance measures can often lead to erroneous results.In this abstract, we propose a method to parcellate whole-brain tractograms efficiently, catering to scenarios where the tractograms cannot be clearly separated owing to inter-subject differences. This is achieved via tract dictionary learning (TractDL) to learn tractogram exemplars for fast, automated analysis of large datasets. Using the dictionaries, whole brain bilateral clustering is applied for simultaneous bundle parcellation in both hemispheres.
Methods
Training: We parameterize and map each streamline to a Hilbert space4. Parcellation is performed in the cosine representation space via a sparse framework with bundle-specific dictionaries, which are learned jointly across bundles. Let $$$\mathbf{X}_t$$$ represent the set of cosine coefficients of the streamlines of $$$t$$$-th bundle.We learn a dictionary $$$\mathbf{D}_t$$$ for the $$$t$$$-th bundle using a sparse dictionary learning framework. Each column of $$$\mathbf{X}_t$$$ can be represented as a linear combination of the columns of an over-complete dictionary $$$\mathbf{D}_t$$$ with sparse coefficients encoded in $$$\mathbf{A}_t$$$. Based on the coefficients, the streamlines can be classified into different bundles. We exploit structured incoherence5 and shared features across bundles so that within-bundle variability is small whereas between-bundle variability is large. To obtain incoherent discriminative dictionaries, we let $$$\mathbf{X} = \left[\mathbf{X}_1,\mathbf{X}_2,...\right]$$$ and $$$\mathbf{D}=\left[\mathbf{D}_1,\mathbf{D}_2,...\right]$$$ and solve the following problem:\begin{equation}\min_{\mathbf{A},\mathbf{D}}\frac{1}{2}\left\| \mathbf{X}-\mathbf{D}\mathbf{A} \right\|_F^2 + \lambda\left\| \mathbf{A} \right\|_1 + \eta \sum_{t_1 \ne t_2} \left\| \mathbf{D}_{t_1}^{\top}\mathbf{D}_{t_2} \right\|_F^2\quad\text{s.t.}\quad\mathbf{A} \succeq 0,\end{equation}where $$$\sum_{t_1 \ne t_2} \left\| \mathbf{D}_{t_1}^{\top}\mathbf{D}_{t_2} \right\|_F^2$$$encourages orthogonality between each dictionary pair6. $$$\lambda$$$ and $$$\eta$$$ are scalar tuning parameters. The size of each bundle dictionary is fixed. Each row of $$$\mathbf{X}$$$ is normalized to have a mean of zero and variance of one to remove the effects of distribution location and scale.Testing: To rapidly classify whole-brain tractograms into bundles, we first perform unsupervised clustering on the tractograms and then label the resulting clusters. To improve clustering robustness, whole-brain bilateral clustering is applied for simultaneous bundle parcellation in both hemispheres. We employ a fast k-medoids clustering algorithm. We randomly select 50 streamlines from each cluster and label each of them via solving the classifier below: \begin{equation}{t}^{*} = \arg\min_{t} \frac{1}{2}\left\| {\mathbf{x}} - \mathbf{D}_t \mathbf{a}_t \right\|_2^2 + \gamma \left\| \mathbf{a}_t \right\|_1 + (1-\gamma)\left\| \mathbf{a}_t \right\|_2, ~\text{s.t.} ~ \mathbf{a}_t \succeq 0,\label{eq:class}\end{equation}where $$${t}^{*}$$$ is the label of streamline $$$\mathbf{x}$$$, and $$$\gamma$$$ is a regularization parameter. The sparse code $$$\mathbf{a}_{t}$$$ of a streamline $$$\mathbf{x}$$$ (cosine representation) with respect to the dictionary of bundle $$$t$$$ can be used for streamline classification. The overall bundle label of each cluster is the label with count fraction (out of 50) greater than a threshold $$$v$$$.
Materials
We evaluated our method using adult and infant data. Adult: The dataset7 used for evaluation contained 72 WM tract bundles for each of 105 young adults from the Human Connectome Project (HCP)8. 70 subjects were used for training and the remaining 35 subjects were used for testing. The fiber streamlines of each subject were transformed to a common space via affine transformation determined by volumetric registration of T1-weighted images. Infant: Multi-shell dMRI data from the Baby Connectome Project (BCP)9.Results
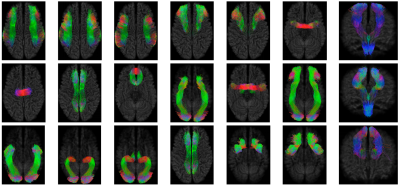
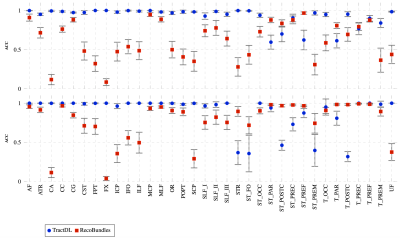
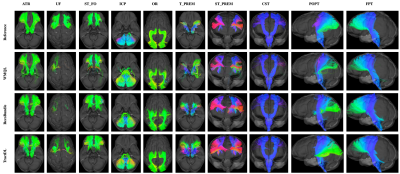
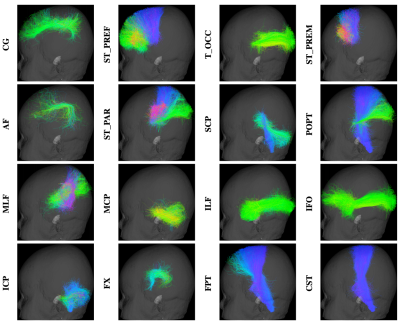
Fig. 1 shows the fiber clustering results by leveraging bilateral symmetry. Fig. 2 reveals that TractDL yields in general higher classification accuracy (ACC) than RecoBundles2, especially in the identification of association and projection tracts. To evaluate the robustness of TractDL, we regenerated the whole brain tractograms using Mrtrix310 (iFOD2 with default parameters) of randomly chosen subject from the testing dataset. Fig. 3 shows that, by employing TractDL, we were able to identify difficult bundles such as AF, CG, CST, FPT, and ILF. To evaluate the generalizability of TractDL, we applied it to a 54-day-old infant, with dMRI data acquired with a protocol that is different from the training HCP subjects. We regenerated the tractograms by employing asymmetry spectrum imaging (ASI) tractography11. Fig. 4 indicates that TractDL can effectively identify the bundles in the infant brain.Conclusion
We have presented a method for fast and robust identification of fiber bundles of interest from a large number of streamlines. The effectiveness, robustness, and generalizability of our method are confirmed by the experimental results.Acknowledgements
This work was supported in part by NIH grants (NS093842 and 1U01MH110274) and the efforts of the UNC/UMN Baby Connectome Project Consortium.References
1. Johansen-Berg H, Behrens TE, editors. Diffusion MRI: from quantitative measurement to in vivo neuroanatomy. Academic Press; 2013.
2. Garyfallidis E, Côté MA, Rheault F, Sidhu J, Hau J, Petit L, Fortin D, Cunanne S, Descoteaux M. Recognition of white matter bundles using local and global streamline-based registration and clustering. NeuroImage. 2018;170:283-95.
3. Zhang F, Wu Y, Norton I, Rigolo L, Rathi Y, Makris N, O'Donnell LJ. An anatomically curated fiber clustering white matter atlas for consistent white matter tract parcellation across the lifespan. NeuroImage. 2018;179:429-47.
4. Chung MK, Adluru N, Lee JE, Lazar M, Lainhart JE, Alexander AL. Cosine series representation of 3D curves and its application to white matter fiber bundles in diffusion tensor imaging. Statistics and its interface. 2010;3(1):69.
5. Fillard P, Descoteaux M, Goh A, Gouttard S, Jeurissen B, Malcolm J, Ramirez-Manzanares A, Reisert M, Sakaie K, Tensaouti F, Yo T. Quantitative evaluation of 10 tractography algorithms on a realistic diffusion MR phantom. Neuroimage. 2011;56(1):220-34.
6. Ramirez I, Sprechmann P, Sapiro G. Classification and clustering via dictionary learning with structured incoherence and shared features. In 2010 IEEE Computer Society Conference on Computer Vision and Pattern Recognition 2010 (pp. 3501-3508). IEEE.
7. Wasserthal J, Neher P, Maier-Hein KH. Tractseg-fast and accurate white matter tract segmentation. NeuroImage. 2018;183:239-53.
8. Van Essen DC, Smith SM, Barch DM, Behrens TE, Yacoub E, Ugurbil K, Wu-Minn HCP Consortium. The WU-Minn human connectome project: an overview. Neuroimage. 2013;80:62-79.
9. Howell BR, Styner MA, Gao W, Yap PT, Wang L, Baluyot K, Yacoub E, Chen G, Potts T, Salzwedel A, Li G. The UNC/UMN baby connectome project (BCP): an overview of the study design and protocol development. NeuroImage. 2019;185:891-905.
10. Tournier JD, Smith R, Raffelt D, Tabbara R, Dhollander T, Pietsch M, Christiaens D, Jeurissen B, Yeh CH, Connelly A. MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. NeuroImage. 2019:116137.
11. Wu Y, Lin W, Shen D, Yap PT, UNC/UMN Baby Connectome Project Consortium. Asymmetry Spectrum Imaging for Baby Diffusion Tractography. InInternational Conference on Information Processing in Medical Imaging 2019 (pp. 319-331). Springer, Cham.
12. Wassermann D, Makris N, Rathi Y, Shenton M, Kikinis R, Kubicki M, Westin CF. The white matter query language: a novel approach for describing human white matter anatomy. Brain Structure and Function. 2016;221(9):4705-21.
Figures