0849
The IronTract challenge: Validation and optimal tractography methods for the HCP diffusion acquisition scheme1Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital and Harvard Medical School, Charlestown, MA, United States, 2Radiology Department, Centre Hospitalier Universitaire Vaudois and University of Lausanne, Lausanne, Switzerland, 3Signal Processing Lab (LTS5), École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland, 4Institute of Imaging Science, Vanderbilt University, Nashville, TN, United States, 5University of Wisconsin, Madison, WI, United States, 6Department of Neuroscience and Biomedical Engineering, Aalto University, Helsinki, Finland, 7Department of Biomedical Engineering, Faculty of Engineering, Yeditepe University, Instanbul, Turkey, 8Department of Neurological Surgery, University of Pittsburgh, Pittsburgh, PA, United States, 9Department of Neuroscience, Brighton and Sussex Medical School, University of Sussex, Brighton, United Kingdom, 10NeuroPoly Lab, Polytechnique Montreal, Montreal, QC, Canada, 11Department of Radiology and BRIC, University of North Carolina, Chapel Hill, NC, United States, 12Neuroscience Research Center, University Magna Graecia of Catanzaro, Catanzaro, Italy, 13CNRS/ISC, Bron, France, 14Université de Bordeaux, Bordeaux, France, 15CNRS/INCIA, Bordeaux, France, 16Center for Advanced Imaging Innovation and Research (CAI2 R), NYU School of Medicine, New York, NY, United States, 17Center for Biomedical Imaging, Dept. of Radiology, NYU School of Medicine, New York, NY, United States, 18Boston Children's Hospital, Boston, MA, United States, 19Montreal Neurological Institute, McGill University, Montreal, QC, Canada, 20Hotchkiss Brain Institute and Department of Radiology, University of Calgary, Calgary, AB, Canada, 21Department of Electrical Engineering, Vanderbilt University, Nashville, TN, United States, 22FIDMAG Germanes Hospitalàries, Sant Boi de Llobregat, Barcelona, Spain, 23Mental Health Research Networking Center (CIBERSAM), Madrid, Spain, 24Translational Imaging in Neurology (ThINK), Department of Medicine and Biomedical Engineering, University Hospital and University of Basel, Basel, Switzerland, 25ICTEAM Institute, Université Catholique de Louvain, Louvain-la-Neuve, Belgium, 26Computer Science Department, University of Verona, Verona, Italy, 27Wellesley College, Wellesley, Wellesley, MA, United States, 28DeepHealth, Inc., Cambridge, MA, United States, 29QMENTA, Inc., Barcelona, Spain, 30Department of Pharmacology and Physiology, University of Rochester School of Medicine, Rochester, NY, United States
Synopsis
We present results from IronTract, the first challenge to evaluate tractography on the two-shell diffusion scheme of the Human Connectome Project (HCP). Accuracy was evaluated by comparison to tracer injections in the same macaque brains as the diffusion data. Training and validation datasets involved different injection sites. We observed that optimizing data analysis with respect to one injection site does not guarantee optimality for another; encouragingly, two teams could achieve consistently high performance in both datasets. We also found that, when analysis methods are optimized, the HCP scheme may achieve similar accuracy as a more demanding diffusion spectrum imaging acquisition.
Introduction
The error-prone nature of diffusion MRI (dMRI) tractography has received considerable attention in recent years, in great part due to tractography challenges that have increased our awareness of the limitations of this technique1-7. Prior challenges, however, used dMRI data that had been either synthesized or acquired with a single, low b-value. This precluded the use of state-of-the-art analysis methods that require multi-shell or Cartesian sampling schemes. Furthermore, it is not clear whether the conclusions of those studies are applicable to the multi-shell, high-angular-resolution dMRI data that are now widely available thanks to large-scale initiatives like the Human Connectome Project (HCP). The IronTract challenge seeks to address this gap by investigating i) which data processing strategies lead to optimal tractography accuracy for the two-shell dMRI acquisition scheme of the lifespan and disease HCP, and ii) whether those methods could achieve even higher accuracy with a different acquisition scheme. Here we present initial results of the challenge and discuss next steps.Methods
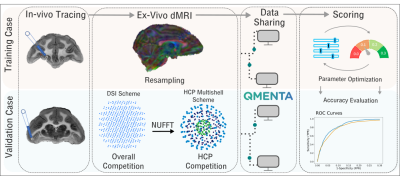
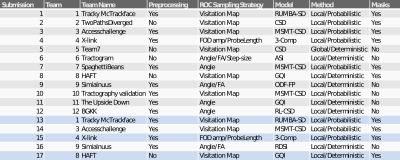
The training and validation cases are part of a previously described dataset that consists of in-vivo tracing and ex-vivo dMRI acquired in the same macaque brains8-10. Tracer data: Bidirectional tracers were injected as previously described11. The training and validation cases consisted of two different brains each of which received a single injection, in the anterior frontal and ventrolateral prefrontal cortex respectively. dMRI data: After fixation, the brains were scanned in a small-bore 4.7T Bruker scanner using 3D EPI, (0.7x0.7x0.7mm, TR=750ms, TE=43ms, 𝛿=15ms, Δ=19ms, maximum b=40,000s/mm2), with 515 volumes corresponding to a Cartesian lattice in q-space. These data were resampled on q-space shells, using a fast implementation of the non-uniform fast Fourier transform (NUFFT)12. We generated data on the two q-shells of the HCP lifespan acquisition scheme (b=1500/ 3000s/mm2, multiplied here by the 4x factor required to achieve comparable diffusion contrast ex-vivo as in-vivo13). Challenge: The challenge was administered through the QMENTA platform (qmenta.com/irontract-challenge/). Participants were blind to the tracer data. For the training case, they uploaded their tractography results and received a score (see below) and ranking. They could repeat this any number of times while they fine tuned the free parameters of their methods to optimize their score. They then applied their optimized analysis pipelines to the validation case, which was used as the basis for the final ranking (Figure 1). Figure of merit: In contrast to prior challenges, participants were asked to upload tractography volumes obtained with multiple thresholds. The thresholding strategy (e.g., angle or probability-based) was left up to the participants. For each tractography volume, true and false positive rates were computed by voxel-wise comparison to the tracer data. The score was the area under the curve (AUC). It was computed for false positive rates in [0,0.3], hence the maximum score was 0.3. We separated the rankings into: i) Overall/DSI: participants were allowed to use any sampling scheme ii) HCP: participants were restricted to the HCP-like, two-shell scheme.Results
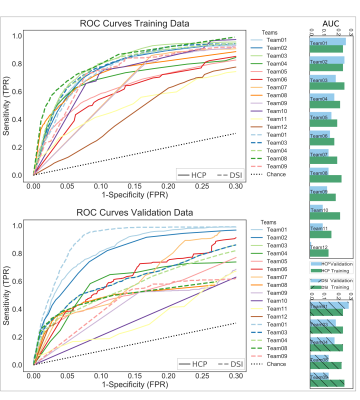
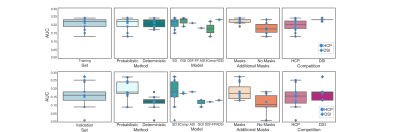
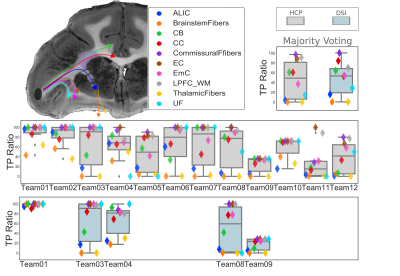
We report results submitted before the MICCAI 2019 conference. Out of 30 registered teams, 12 completed the challenge. There were 227 total submissions (training: 187, validation: 39) and 17 final submissions that were ranked. The diffusion reconstruction and tractography algorithms used are reported in Table 1. Overall, better performance was achieved for the training (mean AUC=0.20) than the validation case (mean AUC=0.15) (Figure 2). Higher AUC scores were obtained using the DSI scheme, probabilistic tractography, spherical deconvolution, and additional constraining masks (Figure 3). We localized the true positives and false negatives for each submission in terms of pathways in the validation case (Figure 4). At a false positive rate=0.1, the sensitivity was variable across different pathways and overall low (HCP=0.57, DSI=0.56). Almost all submissions label regions close to the injection site correctly, but most fail to reconstruct pathways far from it or that require splitting from the main trajectory (eg. brainstem and thalamic fibers). Majority voting analysis confirms this trend.Discussion and Conclusion
Our results show that, when processing methods are tuned appropriately, it is possible to achieve similar tractography accuracy with the HCP and DSI schemes, even though the latter involves 2.8 times more directions and 3.3 times higher maximum b-value. Thus the HCP scheme represents an advantageous trade-off between accuracy and acquisition time. For many of the pipelines employed here, optimizing the methods with respect to accuracy for one seed/injection region did not guarantee optimal performance for another region. This highlights the importance of using anatomical studies from a variety of regions as guidance for tractography. The two injection sites used here project through similar white-matter pathways but reach those pathways from very different angles. The tracing data reveal complex systems of small bundles that travel within and jump between different pathways14. The present results confirm the limited accuracy of tractography when traveling longer distances and through bottle-neck regions, where fibres align and diverge15. Encouragingly, two teams could achieve consistently high performance in both training and validation datasets. In next steps, we will investigate which of their pre/post-processing and tractography methods led to this robustness. We expect our findings to have implications for analyzing the thousands of datasets acquired with the HCP scheme that will soon be publicly available.Acknowledgements
Data acquisition was supported by the National Institute of Mental Health (R01-MH045573). Additional research support was provided by the National Institute of Biomedical Imaging and Bioengineering (R01-EB021265). Imaging was carried out at the Athinoula A. Martinos Center for Biomedical Imaging at the Massachusetts General Hospital, using resources provided by the Center for Functional Neuroimaging Technologies, P41-EB015896, a P41 Biotechnology Resource Grant, and instrumentation supported by the NIH Shared Instrumentation Grant Program (S10RR016811, S10RR023401, S10RR019307, and S10RR023043).
Additional grants that supported part of this work: NIH grants (NS093842, EB022880, and EB006733).
References
1. Daducci A, Canales-Rodriguez EJ, Descoteaux M, Garyfallidis E, Gur Y, Lin YC, et al. Quantitative comparison of reconstruction methods for intra-voxel fiber recovery from diffusion MRI. IEEE transactions on medical imaging. 2014;33(2):384-99.
2. Ning L, Laun F, Gur Y, DiBella EV, Deslauriers-Gauthier S, Megherbi T, et al. Sparse Reconstruction Challenge for diffusion MRI: Validation on a physical phantom to determine which acquisition scheme and analysis method to use? Med Image Anal. 2015;26(1):316-31.
3. Cote MA, Girard G, Bore A, Garyfallidis E, Houde JC, Descoteaux M. Tractometer: towards validation of tractography pipelines. Med Image Anal. 2013;17(7):844-57.
4. Neher PF, Laun FB, Stieltjes B, Maier-Hein KH. Fiberfox: facilitating the creation of realistic white matter software phantoms. Magn Reson Med. 2014;72(5):1460-70.
5. Maier-Hein, K.H., Neher, P.F., Houde, J.C., et al. The challenge of mapping the human connectome based on diffusion tractography. Nat. Commun. 2017;8(1349).
6. Nath V, Schilling KG, Parvathaneni P, et al. Tractography Reproducibility Challenge with Empirical Data (TraCED): The 2017 ISMRM Diffusion Study Group Challenge. J Magn Reson Imaging. 2019
7. Schilling KG, Nath V, Hansen C, Parvathaneni P, et al. Limits to anatomical accuracy of diffusion tractography using modern approaches. NeuroImage. 2019; 185:1–11
8. Z. Safadi, G. Grisot, S. Jbabdi, T. Behrens, S. R. Heilbronner, J. Mandeville, A. Versace, M. L. Phillips, A. Yendiki, S. N. Haber, Functional segmentation of the internal capsule: Linking white matter abnormalities to specific connections, Journal of Neuroscience. 2018; 38(8):2106-17.
9.G. Grisot. S. N. Haber, A. Yendiki, Validation of diffusion MRI models and tractography algorithms using chemical tracing, Proc. Intl. Soc. Mag. Res. Med.. 2018:734
10.W. Tang, S. Jbabdi, Z. Zhu, M. Cottaar, G. Grisot, J. Lehman, A. Yendiki, S. N. Haber A connectional hub in the rostral anterior cingulate cortex links areas of emotion and cognitive control, eLife, In Press, 2019.
11. Suzanne Haber. Tracing intrinsic fiber connections in postmortem human brain with WGA-HRP. Journal of Neuroscience Methods. 1988;23(1):15–22.
12. Jeffrey AF and Bradley PS. Nonuniform fast Fourier transforms using min-max interpolation. IEEE Transactions on Signal Processing. 2003;51(2):560–574.
13. Dyrby TB, William FC, Alexander DC, Jelsing J, Garde E, Søgaard LV. An ex vivo imaging pipeline for producing high quality and high-resolution diffusion-weighted imaging datasets. Human Brain Mapping, 32(4):544–563, 2011.
14. Lehman JF, Greenberg BD, McIntyre CC, Rasmussen SA, Haber SN. Rules ventral prefrontal cortical axons use to reach their targets: implications for diffusion tensor imaging tractography and deep brain stimulation for psychiatric illness. Journal of neuroscience. 2011;31:10392–10402.
15. Aydogan DB, Jacobs R, Dulawa S, Thompson SL, Francois MC, Toga AW, Dong H, Knowles JA, Shi Y. When tractography meets tracer injections: a systematic study of trends and variation sources of diffusion-based connectivity. Brain Struct. Funct. 2018;223: 2841–2858.
16. Tran G and Yonggang Shi. "Fiber orientation and compartment parameter estimation from multi-shell diffusion imaging." IEEE transactions on medical imaging. 2015;34(11):2320-2332.
17. Wu, Y. et al. Asymmetry spectrum imaging for baby diffusion tractography. In International Conference on Information Processing in Medical Imaging. 319–331 (Springer, 2019).
18. Tournier JD, Calamante F, Connelly A. Robust determination of the fibre orientation distribution in diffusion MRI: non-negativity constrained super-resolved spherical deconvolution. Neuroimage. 2017;35(4):1459-1472.
19. Yeh FC, Wedeen VJ, Tseng WY. Generalized q-sampling imaging. IEEE TMI. 2010;29(9)5.
20. Jeurissen B, Tournier JD, Dhollander T, Connelly A, Sijbers J. Multi-tissue constrained spherical deconvolution for improved analysis of multi-shell diffusion mri data. NeuroImage. 2014;103:411-426, 2014.
21. Dhollander T, Raffelt D, Connelly A. Unsupervised 3-tissue response function estimation from single-shell or multi-shell diffusion MR data without a co-registered T1 image. ISMRM Workshop on Breaking the Barriers of Diffusion MRI. 2016.
22. Baete SH, Cloos MA, Lin YC, Placantonakis DG, Shepherd T, Boada FE. Fingerprinting Orientation Distribution Functions in diffusion MRI detects smaller crossing angles. NeuroImage. 2019;198:231-41.
23. Baete SH, Yutzy S, Boada F. Radial q-space sampling for DSI. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2016;76:769-80.
24 Dell'Acqua F, Scifo P, Rizzo G, Catani M, Simmons A, Scotti G, and Fazio F. A modified damped richardson-lucy algorithm to reduce isotropic background effects in spherical deconvolution. Neuroimage. 2010;49(2):1446-1458.
25. Canales-Rodríguez EJ, Daducci A, Sotiropoulos S, Caruyer E, Aja-Fernández S, Radua J, Yurramendi Mendizabal JM, Iturria-Medina Y, Melie-García L, Alemán-Gómez Y, Thiran JP, Sarró S, Pomarol-Clotet E, Salvador R. Spherical Deconvolution of Multichannel Diffusion MRI Data with Non-Gaussian Noise Models and Spatial Regularization. PLoS One. 2015;10(10):e0138910.
Figures