0781
Stop copying contours from Cine to LGE: multimodal learning with disentangled representations needs zero annotations1School of Engineering, University of Edinburgh, Institute of Digital Communications, Edinburgh, United Kingdom, 2Edinburgh Imaging Facility QMRI, Edinburgh, United Kingdom, 3Centre for Cardiovascular Science, Edinburgh, United Kingdom, 4Cedars Sinai Medical Center, Los Angeles, CA, United States, 5The Alan Turing Institute, London, United Kingdom
Synopsis
We propose a novel deep learning method, Multi-modal Spatial Disentanglement Network (MMSDNet), to segment anatomy in medical images. MMSDNet takes advantage of complementary information provided by multiple sequences of the same patient. Even when trained without annotations, it can segment anatomy (e.g., myocardium) in Late Gadolinium Enhancement (LGE) images, which is essential for assessing myocardial infarction. This is achieved by transferring knowledge from the simultaneously acquired cine-MR data where annotations are easier to be obtained. MMSDNet outperforms classical methods including non-linear registration, and simple copying of contours, as well as the state-of-the-art U-Net model.
Background & Introduction:
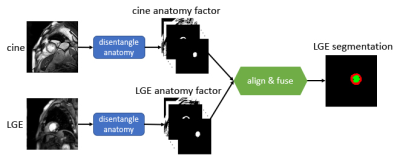
Multiple MR sequences (termed modalities), e.g., cine-MR or Late Gadolinium Enhancement (LGE), are simultaneously acquired to assess organ status. However, for conventional automated analysis, each modality is usually treated in isolation [1]. Successful solutions in one modality cannot be readily translated to another, particularly in deep learning models trained with lots of annotations. Yet, herein arises the opportunity of leveraging complementary information between different modalities. This can be valuable in clinical practice since some pathologies can only be observed in one single modality. The proposed Multi-modal Spatial Disentanglement Network (MMSDNet, schematically depicted in Fig. 1) leverages multi-modal learning to enable full use of information among modalities, which would otherwise be ignored [2]. We argue that such complementary information is crucial in medical image analysis, e.g., segmentation of the left ventricle and the myocardium.Methods:
We introduce the MMSDNet, engaging complementary information provided by multi-modal images. It achieves accurate anatomical segmentation by making full use of the anatomical or pathology information captured by the different modalities.Given 2D cine-MR and LGE images, MMSDNet produces the LGE myocardial and left ventricle segmentation. It is realized by separating information related to anatomy (common in multi-modal images) and image intensities (unique per modality) to achieve a “disentangled” representation with U-Net[4] based anatomy encoders (one per modality) and Variational Auto-encoder based modality encoder. The anatomy factors are binary feature maps with an identical spatial size as the input images, contributing to generating the desired segmentation with a down-stream segmentation network. The modality factor is a latent vector optimized to a Gaussian distribution. Both the anatomy and the modality factors are reconstructing the input images by a self-supervised reconstruction loss with a decoder network.
Input pairing and co-registration: Inputs from different modalities (of the same subject) are not collected simultaneously with different spatial and temporal resolutions. Additionally, cine-MR is dynamic whereas LGE is static. To obtain a mechanism to pair them, a novel automated pairing loss is implemented to correlate these multi-modal data (of the same subject). It compares multiple cine-MR candidate images for a given LGE one. A Multi-layer Perceptron produces weights controlling the contribution of each cine-MR candidate for both the image reconstruction and the segmentation. Additionally, the proposed MMSDNet uses the Spatial Transformer Network [7] (STN) to non-linearly co-register the anatomical factors to be less susceptible to intensity variations attributed to the different sequence characteristics.
Results:
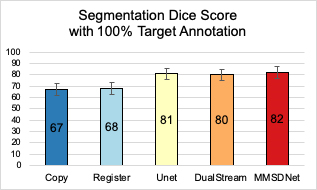
Data: The dataset involved contains cine-MR and LGE images of 28 patients (358 images) with myocardial contour collected at the end-diastolic frame. The spatial resolution is 1.562mm2/pixel (192x192 pixels), and the slice thickness is 9mm.Method for Comparison: We compare our method with state-of-the-art approaches including U-Net[4] and DualStream[5]. A classical approach of copying contours, termed as ‘copy’ (lower bound), is also engaged to evaluate the difference between the ground truth masks of the two modalities. We also include ‘register’ which non-linearly registers the images and then applies the learned transformation to warp the masks. The Dice score is the evaluation metric.
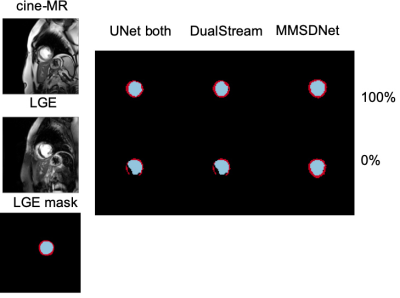
Evaluation: Fig. 2 shows that with full supervision, the mean Dice for MMSDNet, UNet, and DualStream is at similar levels, although the standard deviation of MMSDnet is smaller. Examples are given in Fig. 5.
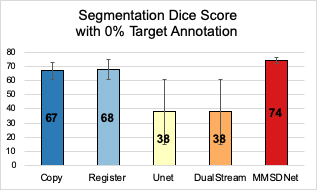
In the scenario (Fig. 3) without is target modality (LGE) annotation, benchmark methods fail to produce accurate segmentation with a high standard deviation. Critically though and visualized in Fig. 5, by simultaneous using both modalities, MMSDNet maintains a good performance. It simultaneously keeps the standard deviation low (at 5%).
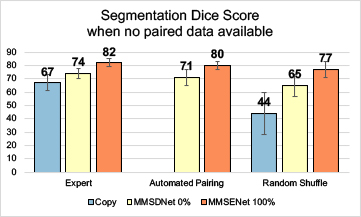
In Fig. 4, the automated pairing is compared with the expert (precisely paired by expert observers) and the random shuffle (randomly shuffle the paired images). The shuffling effect can be seen in the ‘copy’ while performance reduces to 44%. The proposed MMSDNet prevents the performance from dropping drastically by being trained with the proposed automated paring loss. It learns the correlation of the anatomy factors between multi-modal images. Given an input LGE, the new cost automatically selects a corresponding cine-MR slice of the anatomical similarity
Discussion:
The evaluation of the cardiac segmentation with the proposed MMSDNet against state-of-the-art segmentation models has demonstrated the effectiveness obtained by using multiple modalities simultaneously, rather than the traditional isolated treatment. It learns to segment LGE without supervision, transferring knowledge from the co-acquired cine-MR. In addition, the proposed approach can learn how to represent data, align and fuse even when paired data is not available.Conclusion:
Deep learning has achieved considerable performance in several areas of cardiac imaging and particularly in cine-MR. However, this comes at the expense of the necessity of a considerable number of training data. LGE has shown considerable clinical utility in assessing the presence and extent of cardiac infarction but, does require quantification that necessitates myocardial segmentation. Treating LGE segmentation as a learning problem requires lots of annotated data. Instead, the proposed MMSDNet transfers knowledge from the co-acquired cine-MR, achieving competitive results even when only annotations on the cine-MR to train an LGE segmentation model are available. This machine learning driven pipeline is expected to reduce time to clinical practice and translate successes from certain popular (with engineers and computer scientists) modalities to less popular ones.Acknowledgements
This work was supported in part by the US National Institutes of Health (1R01HL136578-01), and used resources provided by the Edinburgh Compute and Data Facility (http://www.ecdf.ed.ac.uk/). S.A. Tsaftaris acknowledges the support of the Royal Academy of Engineering and the Research Chairs and Senior Research Fellowships scheme.References
1. Chartsias, A., Joyce, T., Papanastasiou, G., Semple, S., Williams, M., Newby, D., Dharmakumar, R., Tsaftaris, S.A., 2018. Factorised spatial represen- tation learning: application in semi-supervised myocardial segmentation, in: International Conference on Medical Image Computing and Computer- Assisted Intervention, Springer. pp. 490–498.
2. Kim, H.W., Farzaneh-Far, A., and Kim, R.J.: Cardiovascular magnetic resonance in patients with myocardial infarction: current and emerging applications. JACC 55(1), 1–16 (2009)
3. Valvano, G., Chartsias, A., Leo, A., & Tsaftaris, S. A. (2019). Temporal Consistency Objectives Regularize the Learning of Disentangled Representations. In Domain Adaptation and Representation Transfer and Medical Image Learning with Less Labels and Imperfect Data (pp. 11-19). Springer, Cham. Chicago
4. Olaf Ronneberger, Philipp Fischer, and Thomas Brox. U-net: Convolutional networks for biomedical image segmentation. In MICCAI, pages 234–241. Springer, 2015.
5. Vanya Valindria, Nick Pawlowski, Martin Rajchl, Ioannis Lavdas, Eric Aboagye, Andrea Rockall, Daniel Rueckert, and Ben Glocker. Multi-modal learning from unpaired images: Application to multi-organ segmentation in CT and MRI. In WACV. IEEE, 2018.
6. Colin G Stirrat, Shirjel R Alam, Thomas J MacGillivray, Calum D Gray, Marc R Dweck, Jennifer Raftis, William SA Jenkins, William A Wallace, Renzo Pessotto, Kelvin HH Lim, et al. Ferumoxytol-enhanced magnetic resonance imaging assessing inflammation after myocardial infarction. Heart, 103(19):1528–1535, 2017.
7. Jaderberg, Max, Karen Simonyan, and Andrew Zisserman. "Spatial transformer networks." In Advances in neural information processing systems, pp. 2017-2025. 2015.
Figures