0780
A level-set reformulated as deep recurrent network for left/right ventricle segmentation on cardiac cine MRI1Department of Diagnostic Radiology, The University of Hong Kong, Hong Kong, Hong Kong
Synopsis
We proposed a segmentation method, which is based on a level-set reformulated via a deep recurrent network (RLSNet). This network takes the advantage of U-Net in terms of medical pattern recognition and level-set algorithm in terms of keeping the enclosed and smooth shape of the segmentation contour. We evaluate the network by the segmentation of the left and right ventricles of the heart on cardiac cine Magnetic Resonance Images, which gives greater performance than using U-Net only.
INTRODUCTION
Traditional medical image analysis utilizes level-set based algorithms1,2. This technique iteratively fits an enclosed contour to the boundary of interested objects in the image. And recent attentions focus on using the U-Net network for detection and segmentation. The deep neural network learns non-handcrafted features from a large amount of labeled data via a U-shape network3. It generates a probability map where the pixel intensity, from 0 to 1, indicates the probability of being the desired object or not. The level-set methods provides smooth and enclosed contour while it needs lots of hyper-parameter tuning. U-Net is powerful but it only gives a probability segmentation, where post-processing (e.g. applying a rule of thumb threshold) is still required for accurate segmentation. Therefore, we propose a recurrent level-set network (RLSNet)4, which integrates both level-set mechanism and the U-Net architecture, for the end-to-end segmentation on medical images.METHODS
The proposed network contains two parts, one followed by the other. The first part has the same architecture as U-Net for feature learning. However, instead of using a logistic sigmoid layer at the end producing a probability map, we use a Tanh layer to generate a map with intensity ranges from -1 to 1 (as shown in figure 1a). It can be considered as the energy map which guides the evolution of level-set, as negative value allows moving inward and positive value allows moving outward. The Tanh layer is then connected to the second part of the proposed network, together with a random initial level set Φ0. The second part of the network exploits a gated recurrent network unit to reformulate the level-set evolution (as shown in figure 1b), named recurrent level set network (RLSNet). A gated recurrent network unit contains an updated gate and a reset gate, which determine if an input signal should be added to the previous stored signal or not. The update gate is similar to the regular strategy of solving level-set problems, which iteratively computes the gradient of a predefined level-set energy function and uses gradient descent method until the minimum energy is found. And the reset gate allows the network to learn a flexible stop criterion from the data avoiding lots of testing and tuning. An initial level-set is evolved via cascade RLSNet units which corresponds to the gradient decent procedure in a regular level-set manner (as shown in figure 1c). At last, a cross-entropy loss is used for network training.RESULTS
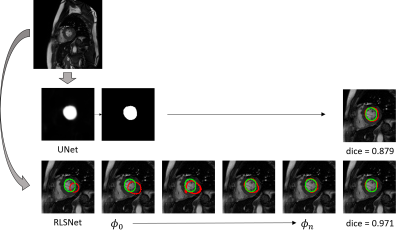
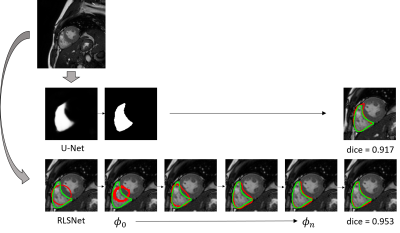
We evaluated the proposed network with cardiac cine MRI images acquired using a 3.0T human scanner (Achieva TX, Philips, Best, Netherlands). We compared the segmentation performance on the left ventricle (LV) and the right ventricle (RV) of the heart with a regular U-Net architecture. The cardiac image dataset contains the short axis (SAX) images of both the end-systolic and end-diastolic phase from 60 subjects. After excluding the image slices of heart base and apex, where the left or the right ventricle can hardly be seen, we obtain 1153 image slices for the left ventricle segmentation and 1155 for the right ventricle. The contour of LV and RV were manually annotated by a cardiac radiologist with level 3 accreditation from our department using the cvi42 software5. We used the data from 40 subjects for training, and the rest 20 for testing. In figure 2, we show the maximum median and average DICE coefficient of the U-Net and the proposed RLSNet using the LV and the RV testing images respectively. Figure 3 shows the difference in segmentation of the two methods on individual LV testing image. It shows that the proposed RLSNet outperforms than using the U-Net only. In figure 4, we give a similar comparison on the right ventricle.DISCUSSION
The proposed RLSNet network reformulates a level-set segmentation in a form of gated recurrent network. It also utilizes the U-Net architecture to reformulate a data-driven energy function for object detection and segmentation. Experimental results on left and right ventricle of the heart on cardiac cine MRI images shows that it outperforms than using U-Net only for segmentation. For future work, we will test the proposed network on a larger set of clinical cine images for further validation.Acknowledgements
The work is supported by the HKU URC Seed Fund.References
1. Soomro, S., Akram, F., Munir, A., Lee, C.H. and Choi, K.N, Segmentation of left and right ventricles in cardiac MRI using active contours. Computational and Mathematical Methods in Medicine, 2017, 2017: 1-16.
2. Li, C., Xu, C., Gui, C. and Fox, M.D., Distance regularized level set evolution and its application to image segmentation. IEEE Transactions on Image Processing, 2010, 19(12): 3243-3254.
3. Ronneberger, O., Fischer, P. & Brox, T. U-net: Convolutional networks for biomedical image segmentation. Lecture Notes in Computer Science (including its subseries Lecture Notes in Artificial Intelligence (LNAI) and Lecture Notes in Bioinformatics (LNBI)), 2015, 9351: 234–241.
4. Le, T.H.N., Quach, K.G., Luu, K., Duong, C.N. and Savvides, M., Reformulating level sets as deep recurrent neural network approach to semantic segmentation. IEEE Transactions on Image Processing, 2018, 27(5): 2393-2407.
5. Circle Cardiovascular Imaging Inc. Calgary, Canada: https://www.circlecvi.com.
Figures