0731
Optimal experimental design in multidimensional diffusion MRI for parameter estimation of biophysical tissue models1Centre for Computational Imaging & Simulation Technologies in Biomedicine (CISTIB), School of Computing and School of Medicine, University of Leeds, Leeds, United Kingdom, 2Radiology, School of Medicine, New York University, New York City, NY, United States, 3Center of Functionally Integrative Neuroscience (CFIN) and MINDLab, Department of Clinical Medicine, Aarhus University, Aarhus, Denmark, 4Department of Physics and Astronomy, Aarhus University, Aarhus, Denmark
Synopsis
It was recently shown that multidimensional diffusion MRI enables well-posed estimation of the Standard Model (SM) for diffusion in white matter. However, various multidimensional acquisitions can achieve this, and there are currently no criteria for efficient data acquisition for SM. We propose an optimal experiment design framework based on Cramér-Rao bounds to maximise accuracy and precision of SM parameter estimation. We explore the high-dimensional continuous acquisition space and identify the optimal combination of b-tensors that minimises estimation error. Simulations and in vivo experiments demonstrate that our optimised acquisition has a reduced estimation error on all SM microstructural parameters.
Introduction
Increased specificity to microstructural changes is the major motivation for developing biophysical models in diffusion MRI (dMRI). For white matter (WM), the Standard Model (SM)1 has been widely adopted. For single pulsed-field gradient dMRI acquisitions, the inverse problem of estimating SM parameters is ill-posed without lengthy acquisitions and extremely high diffusion gradients2,3,4. It has recently been proposed that complementary information from multidimensional dMRI5 resolves this degeneracy6,7. Although several types of multidimensional dMRI acquisitions may be suitable, some will be more efficient and provide more accurate parameter estimates in noisy scenarios.Here we propose a framework for optimal experiment design of multidimensional dMRI and apply it to the SM. The continuous high-dimensional acquisition space of all possible combinations of b-tensor encodings is explored. The optimal acquisition is a nontrivial combination of linear and planar tensor encoding (LTE, PTE), which is corroborated with simulations and in vivo experiments.
Theory
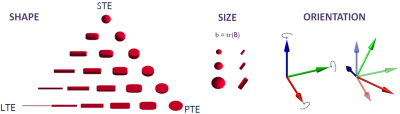
Multidimensional dMRI measurements are represented by a rank-2 symmetric b-tensor $$$\boldsymbol{B}$$$ (Fig.1 shows examples). Tensor eigenvalues define the shape and diffusion weighting (i.e. size), and the eigenvectors define the orientation.The Standard Model signal is the convolution over the unit sphere
$$S(\boldsymbol{B})=S_0\int_{\mathbb{S}^2}\mathcal{P}(\hat{\mathbf{u}})\mathcal{K}(\boldsymbol{B},\hat{\mathbf{u}})\,dS_{\hat{\mathbf{u}}},$$
where $$$S_0\equiv{}S(\boldsymbol{B})|_{\boldsymbol{B}=0}$$$ is the non-weighted diffusion signal, $$$\mathcal{P}(\hat{\mathbf{u}})$$$ the fibres' orientation distribution function (ODF), and
$$\mathcal{K}(\boldsymbol{B},\hat{\mathbf{u}})=f\exp\bigl[-D_{\text{a}}B_{ij}u_iu_j\bigr]+(1-f)\exp\bigl[-bD_\text{e}^\perp-(D_\text{e}^\parallel-D_\text{e}^\perp)B_{ij}u_iu_j\bigr]$$
is the response signal (kernel) from a fibre segment oriented along $$$\hat{\mathbf{u}}$$$, acquired with the b-tensor $$$\boldsymbol{B}$$$ (Einstein's summation notation assumed). $$$f$$$ is the $$$T_2$$$-weighted intra-axonal signal fraction, and $$$D_{\text{a}}$$$, $$$D_\text{e}^\parallel$$$, and $$$D_\text{e}^\perp$$$ the axial intra-axonal, axial extra-axonal and radial extra-axonal diffusivities, respectively.
Cramér-Rao bounds (CRBs) are of widespread use for experiment design in estimation theory8. If some mild regularity conditions are satisfied, CRBs provide a theoretical lower limit to the variance of an unbiased estimator of the model parameters $$$\boldsymbol{\theta}=(\theta_1,\ldots,\theta_N)$$$8:
$$\boldsymbol{\Sigma}_{\boldsymbol{\theta}}\geq\operatorname{CRB}^{\boldsymbol{\theta}}=[I(\boldsymbol{\theta})]^{-1},\quad{}I_{ij}(\boldsymbol{\theta})=-\mathrm{E}\left[\frac{\partial^{2}}{\partial\theta_{i}\partial\theta_{j}}\log{}f(x\,|\,\boldsymbol{\theta})\right],\quad{}i,j=1,\ldots,N,$$
where $$$f(x\,|\,\boldsymbol{\theta})$$$ is the likelihood and $$$I(\boldsymbol{\theta})$$$ the Fisher information.
Methods
We search for the b-tensor set (B-set) maximising the precision in the kernel parameters ($$$\boldsymbol{\psi}=\{f,D_{\text{a}},D_\text{e}^\parallel,D_\text{e}^\perp\}$$$). Thus, our problem separates into the definition of a metric and its optimisation.Metric definition. To simultaneously avoid discrete degeneracy3,4 and maximise precision, we propose a two-step CRB-based metric. First, we compute $$$\operatorname{CRB}^{\boldsymbol{\theta}}$$$, the CRBs for $$$\boldsymbol{\theta}=\{D_{ij},C_{ijk\ell}\}$$$, the tensors from the second-order cumulant expansion of the signal:
$$\log(S)=\log(S_0)-B_{ij}D_{ij}+\tfrac12B_{ij}B_{k\ell}C_{ijk\ell}+{\cal O}(\boldsymbol{B}^3).$$
SM parameters are uniquely determined by $$$\boldsymbol{\theta}$$$ for non-isotropic ODF6,7. Thus, maximising precision on $$$\boldsymbol{\theta}$$$ guarantees non-degenerate kernel estimation. Secondly, we consider the kernel as a function of the cumulants, $$$\boldsymbol{\psi}(\boldsymbol{\theta})$$$, and propagate $$$\operatorname{CRB}^{\boldsymbol{\theta}}\mapsto\operatorname{CRB}^{\boldsymbol{\psi}(\boldsymbol{\theta})}$$$, using the mapping from9:
$$\boldsymbol{\Sigma}_{\boldsymbol{\psi}}\geq\operatorname{CRB}^{\boldsymbol{\psi}}\simeq\frac{\partial\boldsymbol{\psi}(\boldsymbol{\theta})}{\partial\boldsymbol{\theta}}\operatorname{CRB}^{\boldsymbol{\theta}}\frac{\partial\boldsymbol{\psi}(\boldsymbol{\theta})}{\partial\boldsymbol{\theta}}^{t}.$$
Kernel bounds, $$$\operatorname{CRB}^{\boldsymbol{\psi}}$$$, are combined into a scalar and integrated over the (ground truth) kernel and ODF parameter space $$$\cal{H}$$$:
$$F\left(\boldsymbol{\psi},\left\{p_{\ell{}m}\right\}\right)=\sum_{a=1}^{4}\mathrm{CRB}_{aa}^{\boldsymbol{\psi}}/\boldsymbol{\psi}_{a}^{2},\qquad\hat{F}=\frac{1}{\operatorname{vol}(\mathcal{H})}\int_{\mathcal{H}}F\left(\boldsymbol{\psi},\left\{p_{\ell{}m}\right\}\right)d\boldsymbol{\psi}d\boldsymbol{\rho},$$
where $$$\boldsymbol{\rho}=\{p_{\ell{}m}\}$$$ are the ODF spherical harmonics coefficients. To speed up the computation, we average $$$F$$$ over 30 different ODF that correspond to isotropically oriented perpendicular fibre bundles and a single set of kernel parameters ($$$f=0.55,\,D_\text{a}=2.1\mu{}m^2/ms,\,D_\text{e}^{\parallel}=1.8\mu{}m^2/ms,\,D_\text{e}^\perp=0.7\mu{}m^2/ms$$$).
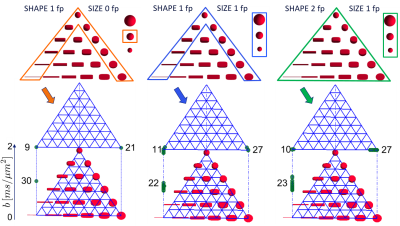
Optimisation. B-tensor eigenvectors were fixed and uniformly distributed over the hemisphere10. The optimisation selected the optimal combination of b-tensor eigenvalues that minimised our metric. We applied three sets of constraints (see Fig.2) progressively increasing the search volume, always limiting maximum b-value to $$$b_{\text{max}}=2ms/\mu{}m^2$$$. B-sets of $$$K\!=\!60$$$ b-tensors were considered. To avoid local minima, stochastic optimisation was used11.
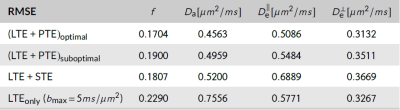
Noise propagation experiments were performed. The optimal protocol was compared against balanced combinations of LTE, PTE and spherical tensor encoding (STE) data (see Imaging). We generated synthetic signals from 7000 random voxels, added noise (SNR=100), estimated the kernel parameters and computed the root mean squared error (RMSE).
Imaging. Three normal volunteers (male-37y/o, male-33y/o, female-32y/o) were imaged on a Siemens Prisma system using a 64-channel head coil. Maxwell-compensated asymmetric waveforms12 were used to generate four protocols, each consisting of a set of b-tensors oriented along 80 isotropically distributed directions and 6 $$$b=0$$$ images. The $$$(\text{LTE+PTE})_\text{optimal}$$$-protocol had $$$30\,\text{LTE}$$$ measurements at $$$b=1ms/\mu{}m^2$$$ and $$$16\,\text{LTE}+34\,\text{PTE}$$$ at $$$b=2ms/\mu{}m^2$$$. The $$$(\text{LTE+PTE})_\text{suboptimal}$$$-protocol had $$$10\,\text{LTE}+10\,\text{PTE}$$$ at $$$b=1ms/\mu{}m^2$$$ and $$$30\,\text{LTE}+30\,\text{PTE}$$$ at $$$b=2ms/\mu{}m^2$$$. The $$$\text{LTE+STE}$$$-protocol had $$$15\,\text{LTE}$$$ at $$$b=1ms/\mu{}m^2$$$, $$$41\,\text{LTE}$$$ at $$$b=2ms/\mu{}m^2$$$ and $$$6\,\text{STE}$$$ at $$$b=0.5,1,1.5,2ms/\mu{}m^2$$$. The $$$\text{LTE}_\text{only}$$$-protocol had $$$20\,\text{LTE}$$$ at $$$b=1ms/\mu{}m^2$$$ and $$$60\,\text{LTE}$$$ at $$$b=2ms/\mu{}m^2$$$. Imaging parameters: TR/E : $$$7200/94ms$$$, resolution: $$$2.5mm$$$ isotropic, in-plane FOV: $$$220mm$$$, GRAPPA acceleration factor $$$2$$$, no partial Fourier. Data was processed using DESIGNER13.
Results
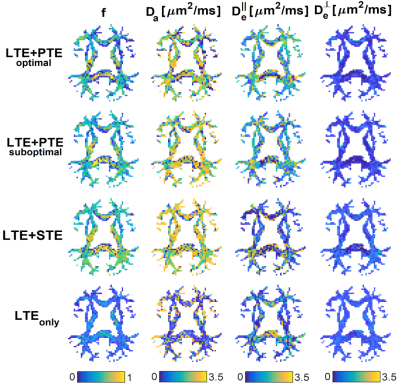
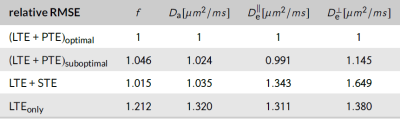
Optimal B-sets converge to two shells of LTE and PTE (see Fig.2). Simulations confirmed that the optimal acquisition has smaller errors than non-informed combinations of LTE+PTE, LTE+STE, or high-b LTE$$$_{\text{only}}$$$ data (see Fig.3 for RMSE values).Fig.4 shows WM parametric maps for one subject. To assess reproducibility, we combined the independent measurements in these acquisitions and estimated SM parameters from the extended dataset. These estimates were used as ground truth and the estimation error was computed for each individual B-set. Averaged across the WM of all subjects, the RMSE was reduced in the optimised LTE+PTE combination (see Fig.5).
Discussion and Conclusion
We for the first time apply a CRB-based optimal experiment design framework to the SM. Our experiments show that a non-trivial combination of LTE and PTE data is optimal with two diffusion weightings.Interestingly, no spherical or non-axially symmetric b-tensors are preferred. Hence, out of all the 2-dimensional b-tensor shape manifold of Fig.1, only 2 discrete shapes are relevant for optimal WM parameter estimation. This may have practical implications for optimised sequence design. Future work will explore high-b and varying TE.
Acknowledgements
This work has been supported by the OCEAN project (EP/M006328/1) and MedIAN Network (EP/N026993/1) both funded by the Engineering and Physical Sciences Research Council (EPSRC) and the European Commission FP7 Project VPH-DARE@IT (FP7-ICT-2011-9-601055). AFF acknowledges support from the Royal Academy of Engineering Chair in Emerging Technologies Scheme (CiET1819\19). SJ acknowledges financial support for a sabbatical at NYU from Aarhus University Research Foundation (AUFF), the Lundbeck Foundation (R291-2017-4375), and Augustinus Fonden (18-1456). SJ is also supported by the Danish National Research Foundation (CFIN), and the Danish Ministry of Science, Innovation, and Education (MINDLab). DSN and EF acknowledge support from NIH under NINDS award R01 NS088040.References
1. Novikov, D. S., Fieremans, E., Jespersen, S. N., and Kiselev, V. G. Quantifying brain microstructure with diffusion MRI: Theory and parameter estimation, NMR in Biomedicine, 2018, 32:e3998.
2. Jelescu, I. O., Veraart, J., Adisetiyo, V., Milla, S. S., Novikov, D. S., and Fieremans, E. One diffusion acquisition and different white matter models: How does microstructure change in human early development based on WMTI and NODDI. NeuroImage, 2015, 107:242-256.
3. Jelescu, I O, Veraart, J., Fieremans, E., and Novikov, D. S. Degeneracy in model parameter estimation for multi-compartmental diffusion in neuronal tissue. NMR in Biomedicine, 2016, 29:33-47.
4. Novikov, D. S., Veraart, J., Jelescu, I. O., and Fieremans, E. Rotationally-invariant mapping of scalar and orientational metrics of neuronal microstructure with diffusion MRI. NeuroImage, 2018, 174:518-538.
5. Topgaard, D. Multidimensional diffusion MRI. Journal of Magnetic Resonance, 2017, 275:98-113.
6. Coelho, S., Pozo, J. M., Jespersen, S. N., Jones, D. K., and Frangi, A. F. Resolving degeneracy in diffusion MRI biophysical model parameter estimation using double diffusion encoding. Magnetic Resonance in Medicine, 2019, 82:395-410.
7. Reisert, M., Kiselev, V. G., and Dhital, B. A unique analytical solution of the white matter standard model using linear and planar encodings. Magnetic Resonance in Medicine, 2019, 81:3819-3825.
8. Kay, S. M. Fundamentals of statistical signal processing: estimation theory. 1993, Englewood Cliffs, N.J.: PTR Prentice-Hall.
9. Pozo, J. M., Coelho, S., and Frangi, A. F. Tensorial formulation allowing to verify or falsify the microstructural standard model from multidimensional diffusion MRI. In Proceedings of the International Society of Magnetic Resonance in Medicine, Wiley, 2019, 3560.
10. Koay, C. G. A simple scheme for generating nearly uniform distribution of antipodally symmetric points on the unit sphere. Journal of Computational Science, 2011, 2(4):377-381.
11. Zelinka, I. Soma | self-organizing migrating algorithm. In New Optimization Techniques in Engineering, 2004, chapter 7, pages 167-217. Springer Berlin Heidelberg.
12. Szczepankiewicz, F., Westin, C.-F., and Nilsson, M. Maxwell-compensated design of asymmetric gradient waveforms for tensor-valued diffusion encoding. Magnetic Resonance in Medicine, 2019, 82(4):1424-1437.
13. Ades-Aron, B., Veraart, J., Kochunov, P., McGuire, S., Sherman, P., Kellner, E., Novikov, D. S., and Fieremans, E. Evaluation of the accuracy and precision of the diffusion parameter estimation with Gibbs and noise removal pipeline. NeuroImage, 2018, 183:532-543.
Figures