0694
Accelerating cardiac hyperpolarized 13C imaging using variational networks for reconstruction
Andreas Dounas1, Valery Vishnevskiy1, Maximilian Fuetterer1, Julia Traechtler1, and Sebastian Kozerke1
1Institute for Biomedical Engineering, University and ETH Zurich, Zurich, Switzerland
1Institute for Biomedical Engineering, University and ETH Zurich, Zurich, Switzerland
Synopsis
A variational network (VN) was implemented and trained on synthetic data to reconstruct multi-echo hyperpolarized 13C data acquired in the in vivo heart. VN reconstruction performance was studied using 2D and 3D synthetic data under low SNR conditions and for acceleration factors of 3 and 9, respectively. Relative to standard gradient descent based reconstruction, the network offers improved reconstruction accuracy and reduced signal leakage between metabolites while preserving information on lactate-to-bicarbonate ratios.
Introduction
In hyperpolarized metabolic imaging of the heart, a trade-off between limited polarization, motion, spatiotemporal resolution, and coverage needs to be found. To this end, various data acquisition strategies have been proposed1. Spatial-spectral excitation of individual resonances with standard image encoding and reconstruction requires compromises in temporal resolution2, hybrid spatial-spectral excitation and multi-echo acquisition3 may suffer from signal spillage between resonances. With standard image reconstruction schemes4 and undersampling, inversion of low signal-to-noise ratio (SNR) signals becomes increasingly ill-posed with resulting image artifacts. However, accelerated imaging is essential to address limited resolution and coverage of hyperpolarized imaging. Variational Networks (VN) have been proposed to implement successive iterative reconstruction steps while learning the necessary filter kernels5 based on in-vivo training data. While VNs were applied for proton-based image reconstruction from undersampled data in high SNR regimes6, their applicability to low SNR conditions and multi-echo data remains to be shown. Moreover, training with synthetic data needs to be addressed as hyperpolarized imaging datasets are scarce. In this work we propose a VN architecture for multi-echo hyperpolarized imaging (VN4DNP). While training is accomplished using synthetic data, the VN can reconstruct undersampled data of the in-vivo heart. Future avenues to accelerate dynamic multi-echo 3D metabolic imaging are explored.Methods
Variational Network:Let $$$\mathbf{E}\in\mathbb{C}^{N_cN_sN_e\times N_pN_m}$$$ be the discretized imaging operator that maps $$$N_m$$$ metabolite images of $$$N_p$$$ pixels each into $$$N_s$$$ k-space samples for $$$N_e$$$ echoes from $$$N_c$$$ coils. We follow5,7 and unroll K=10 iterations of the gradient descent (GD) with $$$n_f$$$ filters per layer:$$\boldsymbol{\rho}^{(k+1)}\leftarrow\boldsymbol{\rho}^{(k)}-\mathbf{E}^\mathrm{H}\psi_{k}(\mathrm{diag}(\mathbf{m})(\mathbf{E}\boldsymbol{\rho}^{(k)}-\mathbf{b}))-\sum_{i\leq n_f}\mathbf{D}^\mathrm{T}_{k,i}\varphi_{k,i}(\mathbf{D}_{k,i}\boldsymbol{\rho}^{(k)} ),$$where $$$\mathbf{m}$$$ is the undersampling mask, $$$\psi$$$ and $$$\varphi$$$ are potential functions parametrized by linear interpolation on a uniform grid with $$$n_i$$$=31 knots. Convolutions $$$\mathbf{D}$$$ are defined by $$$n_c$$$=5x5 kernels with $$$N_m$$$ channels and are constrained to be unit-norm and zero-mean via reparametrization. The reconstructed image is a function of k-space $$$\mathbf{b}$$$ and given by $$$\boldsymbol{\rho}^{(K)}$$$. The network is then defined by $$$\Theta=\{\mathbf{D}_{k,i},\varphi_{k,i},\psi_{k} \}_{k,i}$$$, which is tuned via stochastic optimization over synthetic training data by minimizing the predicted image magnitude error:$$\min_\Theta\mathbb{E}_{\{\boldsymbol{\rho}^\text{gt},\mathbf{b},\mathbf{m}\}\sim\mathcal{T}}\| |\boldsymbol{\rho}^\text{gt}|-|\boldsymbol{\rho}^{(K)}|\|_1$$.
Training Data:
Synthetic training $$$\mathcal{T}$$$ was generated using the XCAT8 model and augmented with random deformations. Normal distributed noise was added for SNR>2. Undersampling was modeled via variable density EPI trajectories. We performed 35$$$\cdot$$$103 iterations of the ADAM9 algorithm with learning rate=10−3 and batch size=5. The general idea of using synthetic data to learn reconstruction of in-vivo data is illustrated in Fig.-1.
In-vivo Data Collection:
In-vivo data was acquired using a dynamic hybrid multi-echo imaging sequence in pigs with inferolateral infarction upon injection of 250mM hyperpolarized [1-13C]pyruvate10. All in-vivo experiments were performed in accordance to the relevant ethics and animal protection legislations. Data was acquired on a 3T MR system using 6 echo-shifted 13C EPI readouts (FOV=220x220mm2, resolution=5x5mm2, slice thickness=20mm, partial Fourier factor (PF)=0.75) distributed across two heartbeats and for multiple dynamic instances. Spectral-spatial excitation attenuated the [1-13C]pyruvate signal while preserving metabolic information of [1-13C]lactate and 13C-bicarbonate. An interleaved constant low-flip angle acquisition was used to yield information on pyruvate concentration.
Image Reconstruction:
To assess reconstruction accuracy of VN4DNP, synthetic test data (FOV=220x220mm2, in-plane resolution=5x5mm2, PF=0.75) with varying SNR and undersampling rates was reconstructed. The results were compared to the conjugate GD-based linear inversion reconstruction. Then, VN4DNP was trained using synthetic 2D data with the same k-space sampling pattern as for in-vivo experiments. Coil sensitivity maps were derived from the pyruvate signal of the interleaved acquisitions11. The trained algorithm was used to reconstruct images from raw in-vivo data to show the generalizability of the network. Subsequently, in-vivo data was retrospectively undersampled by R=3. The network was trained on synthetic undersampled data using the same undersampling masks as for in-vivo undersampling. Finally, undersampling of whole-heart 3D metabolic imaging data (FOV=220x220x110mm3, resolution=5x5x10mm3) was simulated for acceleration factor R=9.
Results
Experiments on synthetic 2D data with varying SNR are illustrated in Fig.-2. Maximum-lactate images are shown with mean lactate and bicarbonate signal over time in the myocardium. The time curves show improved depiction of the signal evolution with VN4DNP compared to standard linear reconstruction across the simulated SNR range. Fig.-3 compares in vivo imaging results of VN4DNP relative to standard reconstruction for PF data and additionally 3-fold undersampled data. The mean signal within the manually segmented myocardium shows improved SNR for VN reconstructions and less signal leakage between metabolites. Axial and coronal cuts of 9-fold undersampled 3D simulations are presented in Fig.-4. Signal time curves are well reproduced down to a peak SNR (pSNR) for lactate of 2.3. At pSNR=2.3, metabolic signals begin to deteriorate.Discussion
In the present work, variational networks for image reconstruction of multi-echo data have successfully been implemented and applied to in-vivo hyperpolarized metabolic imaging data after training in-silico. While the limitations in-silico have been studied, it still has to be shown that required SNRs are achievable in vivo with the proposed acceleration factors.Conclusion
VNs trained on synthetic data allow reconstructing undersampled multi-echo hyperpolarized 13C data with improved performance in low SNR conditions when compared to standard multi-echo reconstruction schemes. The method opens up opportunities to accelerate dynamic multi-echo metabolic imaging protocols.Acknowledgements
No acknowledgement found.References
- Wiesinger F, Weidel E, Menzel MI, et al. IDEAL spiral CSI for dynamic metabolic MR imaging of hyperpolarized [1‐13C]pyruvate. Magn Reson Med. 2012;68(1):8-16.
- Lau AZ, Chen AP, Hurd RE, et al. Spectral–spatial excitation for rapid imaging of DNP compounds. NMR Biomed. 2011;24(8):988-996.
- Sigfridsson A, Weiss K, Wissmann L, et al. Hybrid Multiband Excitation Multiecho Acquisition for Hyperpolarized 13C Spectroscopic Imaging. Magn Reson Med. 2015;73:1713–1717.
- Reeder SB, Pineda AR, Wen Z, et al. Iterative decomposition of water and fat with echo asymmetry and least‐squares estimation (IDEAL): Application with fast spin‐echo imaging. Magn Reson Med. 2005;54(3):636-644.
- Hammernik K, Klatzer T, Kobler E, et al. Learning a variational network for reconstruction of accelerated MRI data. Magn Reson Med. 2018;79(6):3055-71.
- Knoll F, Hammernik K, Kobler E, et al. Assessment of the generalization of learned image reconstruction and the potential for transfer learning. Magn Reson Med. 2019;81(1):116-128.
- Vishnevskiy V, Rau R, Goksel O. Deep Variational Networks with Exponential Weighting for Learning Computed Tomography. MICCAI 2019, LNCS 11769.
- Wissmann L, Santelli C, Segars WP, and Kozerke S. MRXCAT: Realistic numerical phantoms for cardiovascular magnetic resonance. J Cardiov Magn Reson. 2014;16:63.
- Kingma DP, Ba J. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980. 2014.
- Fuetterer M, Traechtler J, Busch J, et al. Longitudinal Hyperpolarized 13C Imaging of Metabolic Changes Following Myocardial Infarction in Pigs. Proc Intl Soc Mag Reson Med. 27 (2019):0721.
- Uecker M, Tamir JI, Ong F, et al. Berkeley Advanced Reconstruction Toolbox (BART). Proc Intl Soc Mag Reson Med. 23 (2015):2486.
Figures
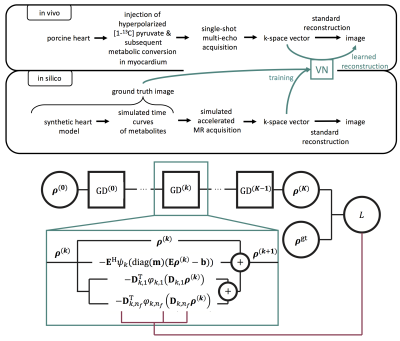
Fig.-1: Top: Pipeline for the VN: In silico data with known ground truth is leveraged to train a model to reconstruct in vivo data. Bottom: VN for loop unrolled GD. Every GD step $$$k$$$ (teal box) uses the output of the previous layer and respective hyperparameters $$$D_{k,i}(k)$$$, $$$\psi_k$$$, and $$$\varphi_{k,i}$$$ to estimate $$$\rho^{(k+1)}$$$. $$$K$$$ GD steps yield an estimate $$$\rho^{(K)}$$$, which can be compared to a ground truth image $$$\rho^{\mathrm{gt}}$$$. The resulting loss $$$L$$$ is then used to update the hyperparameters via backpropagation as shown in (5).

Fig.-2: Reconstruction of time-resolved synthetic 2D data for lactate and bicarbonate. Linear inversion of the full acquisition is compared to the VN with and without threefold incoherent undersampling at different SNRs. The shown images correspond to the time point with maximum lactate signal (teal vertical line). Lactate-to-bicarbonate ratios were calculated for the highlighted areas after subtraction of noise floor. Time curves are scaled equally.
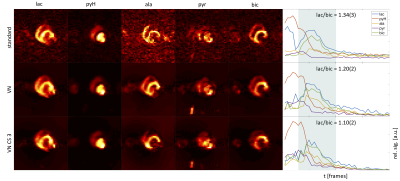
Fig.-3: Reconstruction of in vivo data using a standard conjugate gradient algorithm vs. VN reconstruction of fully and threefold retrospectively undersampled data over all highlighted time frames. Lactate-to-bicarbonate ratios are calculated for the same duration. VN reconstructions show improved noise floor correction and less signal leakage between metabolites as seen for lactate in early time frames.

Fig.-4: Reconstruction of time-resolved synthetic 3D data for lactate and bicarbonate. Results for metabolic signals are shown for ninefold incoherent undersampling within one axial (xy) and one coronal (xz) plane for different SNRs. Metabolic signal is well-depicted with pSNRs of 5.6 for lactate. Lactate-to-bicarbonate ratios are calculated for highlighted time points.