0678
Joint Parallel Imaging reconstruction with Deep Learning for Multi-Contrast Synthetic MRI1Deparment of Electrical & Electronic Engineering, Yonsei Univ., Seoul, Korea, Republic of, 2MR Collaboration and Development, GE Healthcare, Seoul, Korea, Republic of, 3Department of Radiology, Asan medical center, Seoul, Korea, Republic of
Synopsis
Synthetic MRI or magnetic resonance imaging compilation (MAGiC) uses multiple-dynamic multiple-echo acquisition(MDME) and acquires 8 contrast images in a single scan. SENSE or GRAPPA method is conventionally used to reconstruct undersampled acquisition for respective contrast images. However, the method enables limited acceleration up to 2~3. In this study, combined reconstruction method (Joint Parallel Imaging with Deep Learning) is explored. The proposed method shows acceptable image quality with RMSE (4.6%) at the higher acceleration factor (up to 8) comparable to conventional GRAPPA with acceleration rate of 2~3.
Introduction
Synthetic MRI or magnetic resonance imaging compilation (MAGiC) using multiple-dynamic multiple-echo acquisition(MDME) enables the creation of various contrast-weighted images in a single scan.1,2 A typical acquisition method acquires 8 different contrast images with differing time delays and echo times. These images are then used to generate various weighted images such as T1w, T2w, PD, etc. SENSE or GRAPPA method for respective contrast images can be a simple solution to accelerate acquisition time.3,4 However, at the higher acceleration rates (6~8), residual artifacts are unavoidable.In this study, we explored a multi-step reconstruction method, which can utilize joint information between the multi-contrast images to further accelerate the acquisition. In the initial step, a Joint Parallel Imaging method named JVC-GRAPPA(Joint-Virtual Coil GRAPPA) is performed to jointly reconstruction across echoes/contrasts.5 In the following step, remaining residual artifacts are subsequently reduced using Deep Learning(DL). Since the residual artifacts have a particular spatial feature, a data-driven method can enhance the multi-contrast images.
The results show that the proposed method outperforms conventional GRAPPA and JVC-GRAPPA in terms of image quality and RMSE value. Moreover, results with synthesized images (T1w, T2w, PSIR, T2-FLAIR) derived from the accelerated images shows improved image quality.
Method
[Data acquisition]All in-vivo data were acquired on a 3T GE Architect scanner.
Scan protocol for full acquisition : matrix size=300x192, slice thickness=5mm, 25 slices, 48 coils, TR=4000 msec, 4 delays(four automatically calculated saturation) and 2 echo times (TE1=16.4 msec, TE2=90 msec)
[Reconstruction algorithm]
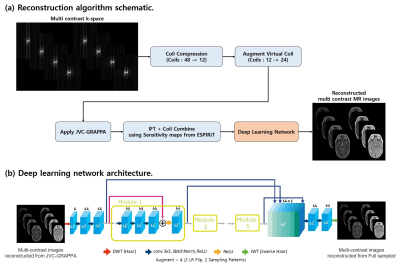
The reconstruction algorithm schematic is shown in Figure 1-a. All experiments used 32 ACS lines, 12 compressed coils using mlSVD(multi linear Singular Value Decomposition).6 Virtual coil augmentation is used to provide additional spatial encoding. Moreover, reconstructed k-space for each coils are combined using coil sensitivity maps estimated from ESPIRiT.7 Finally resulting images were applied to DL network, pretrained on data obtained 9 subjects.
[Deep learning network architecture]
The network architecture for DL is shown in Figure 1-b. For the DL network, WaveResNet8 architecture was used to denoising. This network directly learns the texture information form input MR images using the wavelet frames, leading to better denoising. It substitutes the max-pooling and up-pooling layers in the network by using DWT(Haar Transform) and IWT(inverse Haar) respectively. Data from nine subjects (total 225 slices) were used for training the network. Training data sets were augmented 4 times using Left-Right flip and different sampling patterns. We tested the method on one in-vivo data which was left out from 10 subjects (data from 9 subjects were used for training DL).
[Evaluation]
The resulting images and RMSE values from GRAPPA, JVC-GRAPPA, DL-JVC-GRAPPA were compared. Moreover, results with synthesized images (T1w, T2w, PSIR, T2-FLAIR) derived from the accelerated images were compared.
Results
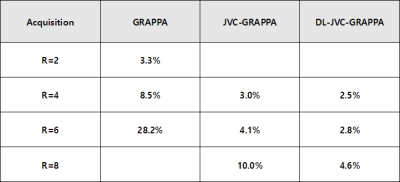
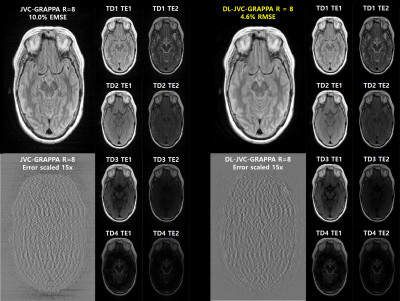
The RMSE values of GRAPPA, JVC-GRAPPA and DL-JVC-GRAPPA are shown in Figure 2. In the situation where the acceleration factor is same, DL-JVC-GRAPPA shows the lowest RMSE value.The reconstructed multi-contrast images with error(scaled 15x) of JVC-GRAPPA and DL-JVC-GRAPPA (R=8) are shown in Figure 3. DL-JVC-GRAPPA provides 2.2x reduction in the RMSE value.
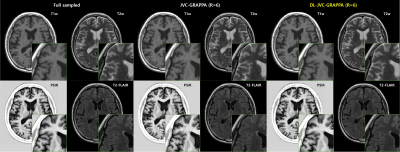
Results with synthesized images(T1w, T2w, PSIR, T2-FLAIR) of Full sampled, JVC-GRAPPA and DL-JVC-GRAPPA (R=6) are shown in Figure 4. As seen, the residual artifacts from JVC-GRAPPA have been reduced on the images reconstructed from DL-JVC-GRAPPA.
Discussion and Conclusion
In this retrospective study, we demonstrated that through Joint Parallel Imaging reconstruction with DL, improved reconstruction results can be achieved. The proposed method shows lower RMSE values than results from JVC-GRAPPA or single contrast GRAPPA. Moreover, the synthesized images (T1w, T2w, PSIR, T2-FLAIR) show similar results. Such reconstruction approach may be beneficial for enabling faster imaging for the synthetic MRI with multi-contrast. We are in the process of modifying the sequence to handle the proposed k-space sampling.Acknowledgements
This work was supported by the National Research Foundation of Korea(NRF) grant funded by the Korea government(MSIT) (NRF-2019R1A2C1090635)References
1. Granberg T, et al. "Clinical feasibility of synthetic MRI in multiple sclerosis: a diagnostic and volumetric validation study." American Journal of Neuroradiology 37.6 (2016): 1023-1029.
2. Krauss W, et al. "Conventional and synthetic MRI in multiple sclerosis: a comparative study." European radiology 28.4 (2018): 1692-1700.
3. Pruessmann KP, et al. "SENSE: sensitivity encoding for fast MRI." Magnetic resonance in medicine 42.5 (1999): 952-962.
4. Griswold MA, et al. "Generalized autocalibrating partially parallel acquisitions (GRAPPA)." Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 47.6 (2002): 1202-1210.
5. Bilgic B, et al. "Improving parallel imaging by jointly reconstructing multi‐contrast data." Magnetic resonance in medicine 80.2 (2018): 619-632.
6. Biyik E, et al. "Reconstruction by calibration over tensors for multi‐coil multi‐acquisition balanced SSFP imaging." Magnetic resonance in medicine 79.5 (2018): 2542-2554.
7. Uecker M, et al. "ESPIRiT—an eigenvalue approach to autocalibrating parallel MRI: where SENSE meets GRAPPA." Magnetic resonance in medicine 71.3 (2014): 990-1001.
8. E. Kang, et al. "Deep convolutional framelet denosing for low-dose CT via wavelet residual network." IEEE transactions on medical imaging 37.6 (2018): 1358-1369.
Figures