0642
Whole-body profiling of early cancer metastasis using multimodality reporter gene imaging1Medical Biophysics, University of Western Ontario, London, ON, Canada, 2Medical Imaging Laboratories, Robarts Research Institute, London, ON, Canada, 3Medical Biophysics, Robarts Research Institute, London, ON, Canada, 4Lawson Health Research Institute, London, ON, Canada
Synopsis
Organic anion-transporting polypeptide 1b3 (Oatp1b3) is a protein derived from the human liver that is capable of taking up Gd-EOB-DTPA, a clinical contrast agent, into cells. We synthetically express the Oatp1b3 gene on breast cancer cells and are able to track them throughout the bodies of preclinical animal models with high sensitivity and resolution as they metastasize. In the future, we hope to develop Oatp1b3 as a tool to track the activation and location of gene and cellular therapies in patients on MRI.
Unlike optical modalities, MRI provides non-invasive, whole-body, non-scattered, high resolution, 3D information with surrounding anatomical context. Several reporter genes for MRI have been previously described2. A member of the organic anion-transporting polypeptide 1 (Oatp1) family, called Oatp1a1, was demonstrated to transport gadolinium ethoxybenzyl diethylenetriaminepentaacetic acid into engineered cells (Gd-EOB-DTPA)3. It provides several desirable characteristics, including positive contrast, contrast reversibility, sensitivity at clinical field strengths, and enhancement patterns on MRI that spatially correlate to the location of viable engineered cells on fluorescent histology4.
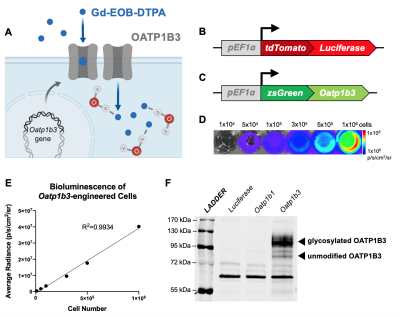
Our own work has recognized that human Oatp1b3 can also take up Gd-EOB-DTPA, allowing for in vivo cell tracking of engineered cells (Figure 1A). The objective of this study was to develop a multimodality reporter system for BLI/MRI, and assess its utility for longitudinal whole-body imaging of spontaneous breast cancer metastases in mice.
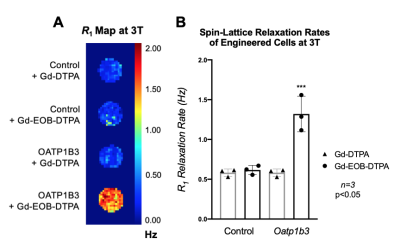
Methods. Triple negative breast cancer cells (MDA-MB-231) were engineered with lentivirus to constitutively co-express the fluorescent protein tdTomato and firefly luciferase for BLI (Figure 1B). A subset then was further engineered to co-express the fluorescent protein zsGreen and Oatp1b3 for MRI (Figure 1C). Luciferase- or Luciferase/Oatp1b3-expressing cells were incubated with either 1.6 mM Gd-EOB-DTPA or Gd-DTPA for 1 hour, washed and pelleted. An inversion recovery experiment at 3 Tesla was performed to determine R1 relaxation rates.
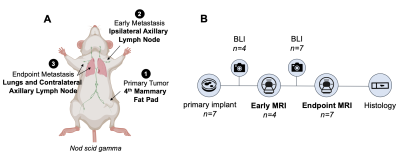
For in vivo imaging, female nod scid gamma mice were orthotopically implanted with 3x105 Luciferase- (n=3) or Luciferase/Oatp1b3-expressing (n=7) cells into the fourth mammary fat pad. Mice were monitored for spontaneous metastases with BLI. High resolution (200 μm3) T1-weighted images were acquired at 3 Tesla both before and 5 hours after administration of 0.5 mmol/kg Gd-EOB-DTPA. Metastatic lesions were manually segmented on pre- and post-contrast images to delineate volumes-of-interest.
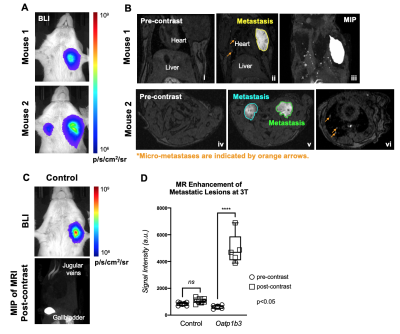
Results. Breast cancer cells were successfully engineered and sorted for tdTomato and zsGreen fluorescence. Average bioluminescent radiance (p/s/cm2/sr) exhibited a strong positive correlation to in vitro cell number (R2=0.9934) (Figure 1D-E). Western blotting confirmed expression of the Oatp1b3 transporter in Luciferase/Oatp1b3-engineered cells (Figure 1F). Luciferase/Oatp1b3-expressing cells treated with Gd-EOB-DTPA exhibited significantly increased spin-lattice relaxation rates (1.320±0.2229 Hz) relative to all other controls (n=3, p=0.0003) (Figure 2). At endpoint MRI, macro-metastases of mice burdened with Luciferase/Oatp1b3-expressing primary tumours (n=3) exhibited significantly increased signal intensity on post-contrast T1-weighted images (4926±1159 a.u.) relative to pre-contrast images (617.5±140.8 a.u.), as well as to macro-metastases of mice burdened with control Luciferase-expressing primary tumours (n=3) both pre-contrast (840.1±122.2 a.u.) and post-contrast (1058±198.0 a.u.) (Figure 4).
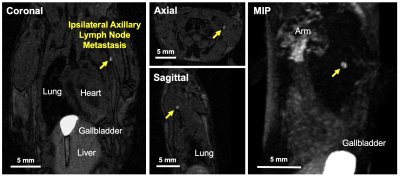
Interestingly, small foci of contrast enhancement were observed on post-contrast MRI within the lungs of mice burdened with Luciferase/Oatp1b3-expressing primary tumours (Figure 4B: ii, iii, vi). On BLI, these foci were obscured by larger macro-metastases (Figure 4A), and were not observed on post-contrast MRI of control animals (Figure 4C), suggesting that small numbers of cells could be sensitively detected with Oatp1b3. With a second cohort of animals (n=4), we detected early axillary lymph node metastasis (≤1mm3) just 10 days following primary tumour implantation, and within 48 hours of the first metastatic detection on BLI (Figure 5, yellow arrow).
Discussion. Maximizing sensitivity is key to improving our ability to visualize and quantify cancer cell metastasis in vivo. While BLI provides sensitive whole-body information on locations of engineered cells, smaller tumours may go undetected due to light scattering from larger lesions. The Oatp1b3 reporter gene can mitigate the limitations of BLI for tracking metastatic disease. Here, we demonstrate that cancer cells could be engineered to express Oatp1b3 and retain reporter gene activity once they acquire a migratory phenotype, allowing for whole-body detection of engineered cells with sensitivity. We show that Oatp1b3 can be used to track the metastatic process at the early stage (single lymph node) and at later stages once the cancer has spread to multiple lymph nodes and other organs e.g. lungs. Importantly, MR reporter gene development has largely focused on iron-based negative contrast, which already exists within living organisms in critical regions such as the lungs.
Oatp1b3 MRI shows paves the path towards molecular imaging of reporter gene-expressing cells with combined high resolution, sensitivity and 3D spatial information with surrounding anatomical context. Future work focuses on completing the cohort of animals for early-stage metastatic detection, and characterizing Gd-EOB-DTPA uptake by Oatp1b3-expressing cells in vivo with kinetic modeling. In the long term, we hope to extend Oatp1b3 utility towards becoming the first clinical-grade MRI reporter for tracking of gene and cellular therapies in patients.
Acknowledgements
No acknowledgement found.References
1. Kubota SI, Takahashi K, Nishida J ... Ueda HR (2017). Whole-Body Profiling of Cancer Metastasis with Single-Cell Resolution. Cell Rep 20(1): 236-250.
2. Yang C, Tian R, Liu T, Liu G (2016). MRI Reporter Genes for Noninvasive Molecular Imaging. Molecules 21(5): 580.
3. Patrick PS, Hammersleya J, Loizoua L ... Brindle KM (2013). Dual-modality gene reporter for in vivo imaging. PNAS 111(1): 415-20.
4. Nyström NN, Hamilton AM, Xia W ... Ronald JA (2019). Longitudinal Visualization of Viable Cancer Cell Intratumoral Distribution in Mouse Models Using Oatp1a1-Enhanced Magnetic Resonance Imaging. Invest Radiol 54(5): 302-311.
Figures