0579
Learning-based approach for accelerated IVIM imaging in the placenta1Key Laboratory for Biomedical Engineering of Ministry of Education, Department of Biomedical Engineering, College of Biomedical Engineering & Instrument Science, Zhejiang University, Hangzhou, China, 2Department of Medical Imaging and Radiological Sciences, Chang Gung University, Taoyuan, Taiwan, 3Department of Radiology,Women's Hospital,School of Medicine, Zhejiang University, Hangzhou, China, 4Purple-river software corporation, Shenzhen, China
Synopsis
Q-space learning has shown its potential in accelerating Q-space sampling in diffusion MRI. This study proposed a new deep learning framework to accelerate intravoxel incoherent motion (IVIM) imaging and to estimate IVIM parameters from a small number of b values in the human placenta. The results demonstrated the feasibility of a reduced IVIM protocol using the proposed framework, which may help to accelerate the acquisition and reduce motion for placental IVIM.
Introduction
Intravoxel incoherent motion (IVIM) has shown to be a useful tool in assessing the microcirculatory flow in the placenta [1, 2], with potential values in the diagnosis of abnormal placental function1-2. However, the conventional IVIM model fitting usually requires acquisition of a series of b-values, leading to relatively long acquisition time and motion artifacts. This becomes a particular problem for placental IVIM, as the patients are less tolerant and the images are more susceptible to motion. Here, we proposed a Q-space learning approach [3], aiming to achieve accurate IVIM estimation with fewer b-values.Materials and Methods
Data acquisition: Diffusion-weighted imaging (DWI) with the whole uterus coverage was acquired from nine normal pregnant women with gestational ages from 28 weeks to 36 weeks, with IRB approval and written consent. The scans were performed on a 1.5T GE SIGNA HDXT scanner, with the following imaging parameters: single-shot EPI, TR = 3000ms, TE = 76ms, 1.25 × 1.25 mm2 in-plane resolution, matrix size = 256 × 256, field of view = 320 × 320 mm2, 15 slices with a slice thickness of 4 mm. Diffusion-weighted gradients were applied in three directions using b-values of 0, 10, 20, 50, 80, 100, 150, 200, 300, 500 and 800s/mm2.IVIM fitting: The IVIM data first underwent motion correction by linear registration of the images to the mean DWI, which was performed in FSL (FMRIB Software Library, University of Oxford, UK). This procedure was repeated to obtain an optimal mean DWI, which turned out to be important for the registration accuracy. Model fitting was performed according to the bi-exponential IVIM model using a segmented approach [4-6] in MATLAB. The model fitting results were defined as ground truth and used as training and test labels in all experiments.
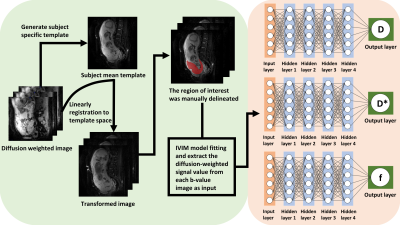
Q-space learning: The structure of the multilayer network was presented in Fig. 1. The network consists of one input layer, four hidden layers and one output layer. The four hidden layers are fully connected, and the number of neurons is equal to the number of features entered. In this deep learning framework, each image voxel serves as an input sample. The input is made of a vector of b-values and the normalized diffusion-weighted signal S(b)/S0 at each b-value. IVIM parameters (tissue water diffusion coefficient (D), pseudo-diffusion coefficient (D*) and perfusion fraction (f)) are the output of multilayer perceptron. The three target parameters were separated and each has its own network. An exponential linear unit (ELU) activation function is added after each fully connected layer. This proposed approach adjusts the parameters of the multilayer network so that the output of the network is forced to match the target metric. The cost function, which is defined as the mean square error between the ground truth and the network’s output, is minimized by using an Adam optimizer [7]. The network is uniformly cycled n times until the network converges. The method was trained on six subjects (approximately 136330 voxels) with reduced b-values (3 b-values at or 5 b-values), and tested on three independent subjects to test its performance.
Results
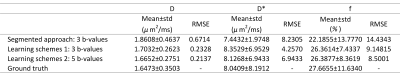
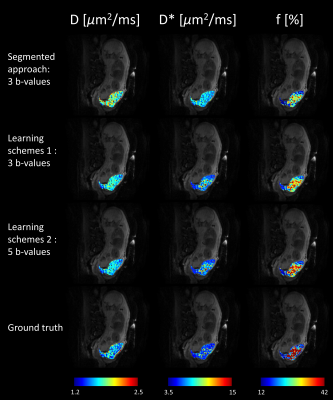
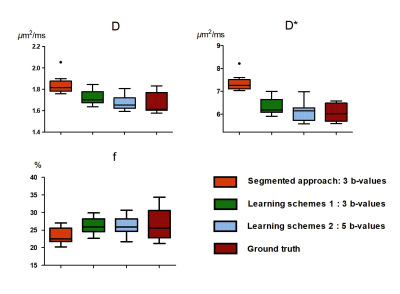
Table 1 shows the performance of IVIM estimation by the proposed network at 3 b-values (b=50, 150, 500s/mm2), 5 b-values (b =20, 80, 150, 300, 500s/mm2), in comparison with the ground truth and a previously proposed simplified approach [8]. Fig. 2 displays maps of the three IVIM parameters estimated by the two proposed schemes (3 and 5 b-values) in the training dataset and testing dataset, in comparison with the simplified 3-point fitting method and the ground truth. Fig. 3 demonstrates the boxplots of intravoxel incoherent motion parameters (D, f, and D*) fitted in placenta by the proposed network, in comparison with the 3-point fitting method and the ground truth. It is noticeable that the result estimated by two proposed schemes were both close to ground truth in D and D*, but differences were observed in f compared with ground truth. Scheme 2 (5 b-values input) showed a better performance than scheme 1(3 b-values input).Discussion and Conclusion
Our results demonstrated the feasibility of using Q-space learning approach to accelerate IVIM acquisition. The results indicated that the majority of the information in the IVIM curve could be recovered from a limited subset of DWIs in the placenta. The choice of b-values is critical for accurate estimation. We chose the reflection point (b=150 s/mm2) in the signal decay curve and a low and high b-value (b=50 and 500 s/mm2) to capture the pseudo-diffusion and tissue water diffusion behaviors. The choice of b-values may vary in other applications, e.g., for placentas at different gestational ages or placentas with certain types of disorders. Therefore, the generalizability of the proposed network needs to be investigated, and we may need train a network with larger samples that includes various situations in future work. In addition, the results indicated that estimation accuracy increase with the number of b-values. In practice, one can choose the number of used DWI and achieve a better trade-off between scan time and estimation accuracy using a deep learning approach. The proposed approach may facilitate IVIM imaging of the placenta with a minimized acquisition protocol with reduced scan time and less susceptibility to motion.Acknowledgements
This work was supported by the Natural Science Foundation of China (61801424, 81971606, and 91859201) and the Ministry of Science and Technology of the People’s Republic of China (2018YFE0114600).References
1.Slator, P.J., et al., Placenta microstructure and microcirculation imaging with diffusion MRI. Magnetic resonance in medicine, 2018. 80(2): p. 756-766.
2.Vedmedovska, N., et al., Placental pathology in fetal growth restriction. European Journal of Obstetrics & Gynecology and Reproductive Biology, 2011. 155(1): p. 36-40.
3.Golkov, V., et al., Q-space deep learning: twelve-fold shorter and model-free diffusion MRI scans. IEEE transactions on medical imaging, 2016. 35(5): p. 1344-1351.
4.Barbieri, S., et al., Impact of the calculation algorithm on biexponential fitting of diffusion‐weighted MRI in upper abdominal organs. Magnetic resonance in medicine, 2016. 75(5): p. 2175-2184.
5.Merisaari, H., et al., Fitting methods for intravoxel incoherent motion imaging of prostate cancer on region of interest level: Repeatability and gleason score prediction. Magnetic resonance in medicine, 2017. 77(3): p. 1249-1264.
6.Pekar, J., C.T. Moonen, and P.C. van Zijl, On the precision of diffusion/perfusion imaging by gradient sensitization. Magnetic resonance in medicine, 1992. 23(1): p. 122-129.
7.Kingma, D.P. and J. Ba, Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980, 2014.
8.Conklin, J., et al., A simplified model for intravoxel incoherent motion perfusion imaging of the brain. American Journal of Neuroradiology, 2016. 37(12): p. 2251-2257.
Figures