0546
Don’t Lose Your Face - Refacing for Improved Morphometry
Till Huelnhagen1,2,3, Mário João Fartaria1,2,3, Ricardo Corredor-Jerez1,2,3, Mazen Fouad A. Wali Mahdi1, Gian Franco Piredda1,2,3, Bénédicte Maréchal1,2,3, Jonas Richiardi1,2, and Tobias Kober1,2,3
1Advanced Clinical Imaging Technology, Siemens Healthcare AG, Lausanne, Switzerland, Lausanne, Switzerland, 2Department of Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 3LTS5, École Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland
1Advanced Clinical Imaging Technology, Siemens Healthcare AG, Lausanne, Switzerland, Lausanne, Switzerland, 2Department of Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 3LTS5, École Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland
Synopsis
A growing amount of imaging data is made publicly available. While this is desirable for science and its reproducibility, privacy concerns increase. As the shape of a face can be recovered based on MR images, an increased number of studies remove the face from the data to prevent biometric identification. This defacing can, however, pose a challenge to existing post-processing pipelines e.g. brain volume assessment. This work investigates the impact of regenerating facial structures in defaced images on morphometry in a large cohort using a deep neural network. The results show that refacing can prevent volumetric errors induced by defacing.
Purpose
Routine structural brain MRI scans allow 3D face-reconstruction of individuals and thus application of biometric methods for identification. This poses a privacy concern when sharing such data or making them publicly available1, which has led to a demand to de-identify the actual MR image data, typically by defacing or blurring. Consequently, an increasing number of large registries that collect neuro imaging data such as the UK Biobank2 or the Human Connectome Project3 are applying such methods before making scans available to researchers. This can, however, pose a challenge for automated post-processing techniques like brain morphometry or lesion detection that are often designed to have whole-head MR-datasets as input. Using such algorithms with defaced data can cause them to fail or affect e.g. volumetric results4. To address this issue, this work proposes to regenerate facial features (refacing) using a deep neural network with the aim to improve automated processing pipeline results. The feasibility of the approach is examined by investigating its impact on brain volume measurements in a large cohort. Refacing of defaced images has previously been proposed5 but not on whole 3D-volumes, and with the goal to investigate potential privacy issues instead of the impact on automated processing pipelines.Methods
We included 1832 T1-weighted MRI-scans of healthy controls and additional 50 patient scans of the TADPOLE dataset6 from ADNI7 in this study. First, all datasets were reoriented to have ASL image orientation. Subsequently, images were defaced using pydeface (8). A Pytorch implementation of the Pix2Pix 2D-generative adversarial network9,10 was trained to reface defaced images using 50 healthy-control and 50 patient scans. To provide 3D-context information to the network, for each slice, also the two neighboring slices were provided as additional input channels (a 2.5D-approach). The trained model was applied to all defaced volumes in the whole dataset. Refacing took about 5 seconds/case on a GPU (NVIDIA GTX-1080-Ti). To avoid unwanted changes by refacing in areas not affected by defacing, voxels that had not been removed by defacing were replaced in the refaced images by the defaced original. Brain segmentation was performed on original, defaced and refaced images using a prototype software (MorphoBox11), developed to work with whole-head images. Volumetric results were compared for the three scenarios and paired Wilcoxon tests were used to test for significant differences. The workflow is illustrated in Figure 1. To avoid bias, subjects of which there was at least one scan in the training set were excluded from the analysis. Cases for which defacing cut parts of the brain or did not remove the face at all were also excluded (n=103). 164 cases for which the refacing routine did not produce any output where excluded as well, leaving a total of 978 scans for the analysis.Results
Figure 2 shows original, defaced and refaced slices of an example test subject together with corresponding 3D-surface renderings. While the regenerated facial structures are blurred and their shape is not identical to the original, the network was able to generate face-like structures with an image contrast that resembles well the original image. Overall, volumetric results were significantly different compared to the original images in 11 out of 17 brain regions after defacing, compared to only two regions after refacing (Table 1). Volumetric differences introduced by the defacing were mostly recovered by refacing (Figure 3,Table 1). Overall differences in the three scenarios where low regarding the whole cohort, indicating that the segmentation algorithm used is rather robust against defacing, but the algorithm failed in some cases after defacing. Such failures could mostly be recovered after refacing (Figure 3,Figure 4). After defacing, morphometric differences of >5% in at least one structure out of gray matter, white matter, cerebrospinal fluid or total intracranial volume were observed in 67 of 1022 cases (6.56%). After refacing, this number went down to 20 (1.96%). For the 67 cases volumetric results were significantly different in 15 out of 17 regions after defacing compared to the original, while after refacing no region showed significant differences.Discussion and Conclusion
We investigated the impact of regenerating facial structures in defaced images on brain volumetric measurements. The results show that refacing can significantly benefit brain morphometry of defaced data. Brain segmentation errors induced by defacing could mostly be prevented by refacing, resulting in significantly improved volumetric results. The choice of a 2.5D-network was driven by resource constraints, as a 3D-network would have needed much more memory with consequently significantly increased hardware requirements. A 3D-network would be obviously more suited for the task of refacing to take advantage of the whole 3D-context but was not possible to realize at this point. Still, the compromise to provide 3D-context by neighboring slices proved suitable to achieve good 3D-consistency. Employing a 3D-model and expansion of the training set will be considered in future work. While refacing can only be an intermediate step towards algorithms specifically designed to work reliably on defaced image data, the proposed approach can help to overcome issues of well-validated, existing pipelines until such solutions will be widely available and equally well evaluated. The low processing time and limited resources needed to run the network make it also easy to implement into existing processing pipelines.Acknowledgements
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.References
1. Schwarz CG, Kremers WK, Therneau TM, et al. Identification of Anonymous MRI Research Participants with Face-Recognition Software. N Engl J Med 2019;38:1684–1686 doi: 10.1056/NEJMc1908881. 2. Sudlow C, Gallacher J, Allen N, et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLoS Med. 2015 doi: 10.1371/journal.pmed.1001779. 3. Van Essen DC, Smith SM, Barch DM, Behrens TEJ, Yacoub E, Ugurbil K. The WU-Minn Human Connectome Project: An overview. Neuroimage 2013 doi: 10.1016/j.neuroimage.2013.05.041. 4. de Sitter A, Visser M, Brouwer I, et al. Impact of removing facial features from MR images of MS patients on automatic lesion and atrophy metrics. Mult. Scler. J. 2017;23:226. 5. Bischoff-Grethe A, Ozyurt IB, Busa E, et al. A technique for the deidentification of structural brain MR images. Hum. Brain Mapp. 2007 doi: 10.1002/hbm.20312. 6. Poldrack RA. Pydeface. https://github.com/poldracklab/pydeface/. 7. Abramian D, Eklund A. REFACING : RECONSTRUCTING ANONYMIZED FACIAL FEATURES USING GANS. 2019 IEEE 16th Int. Symp. Biomed. Imaging (ISBI 2019) 2018:2–6. 8. TADPOLE, https://tadpole.grand-challenge.org constructed by the EuroPOND consortium http://europond.eu funded by the European Union’s Horizon 2020 research and innovation programme under grant agreement No 666992. 9. Alzheimer’s Disease Neuroimaging Initiative (ADNI). 10. Isola P, Zhu JY, Zhou T, Efros AA. Image-to-image translation with conditional adversarial networks. Proc. - 30th IEEE Conf. Comput. Vis. Pattern Recognition, CVPR 2017 2017;2017-Janua:5967–5976 doi: 10.1109/CVPR.2017.632. 11. Zhu JY, Park T, Isola P, Efros AA. Unpaired Image-to-Image Translation Using Cycle-Consistent Adversarial Networks. In: Proceedings of the IEEE International Conference on Computer Vision. ; 2017. doi: 10.1109/ICCV.2017.244. 12. Schmitter D, Roche A, Maréchal B, et al. An evaluation of volume-based morphometry for prediction of mild cognitive impairment and Alzheimer’s disease. NeuroImage Clin. 2015;7:7–17 doi: 10.1016/j.nicl.2014.11.001.Figures
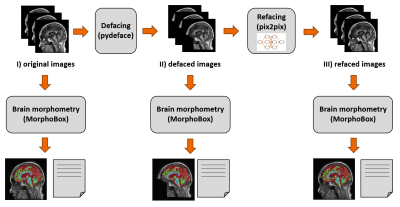
Figure 1: Illustration of the processing scheme. Original T1-weighted
image volumes are defaced and subsequently refaced using the neural network.
Brain segmentation and morphometry is carried out for all three scenarios and
results are compared.

Figure 2: Mid sagittal slices (top) and iso surface renderings (bottom) of an
example test subject for original (left), defaced (center) and refaced (right) images. Regenerated facial structures
are blurred and not perfectly identical to the original face, but their
contrast and overall shape matches well with the original image, which will
e.g. support image registration to a template. The volume rendering shows that
refacing worked consistently in the slice direction.
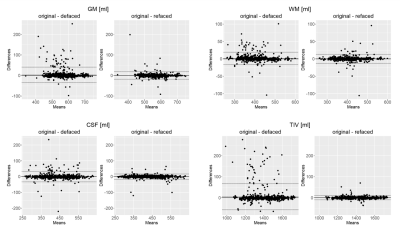
Figure 3: Bland-Altman plots comparing volume differences for
gray matter (GM), white matter (WM), cerebrospinal-fluid (CSF) and total
intracranial volume (TIV) after defacing and refacing against the original. Volumetric
differences occurring after defacing are largely diminished after refacing.
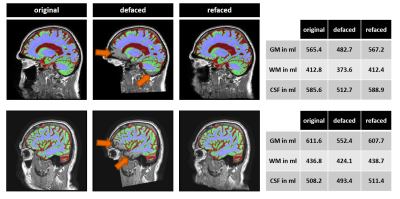
Figure
4: Two
example cases for which refacing significantly improved brain segmentation: Left:
Brain segmentation results overlaid onto corresponding T1-weighted images.
Large segmentation errors (arrows) in the defaced images disappear after refacing.
Volumes of brain structures differ substantially for original and defaced
images but are in the same range as for the original after refacing (right).
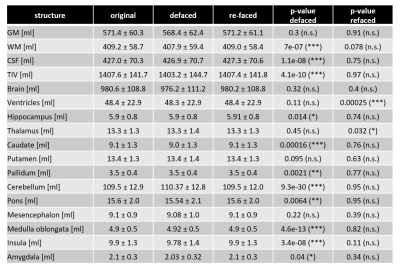
Table 1: Mean volumes and standard deviation of main brain structures
in all datasets for original, defaced and refaced images. Defacing induced
significant differences in several brain regions compared to original
images. After refacing, volumetric differences for all structures except for
ventricles and thalamus were not significant.
The similar means and standard deviations in the defaced images compared to the
original values indicate rather high robustness of the employed segmentation
algorithm against defacing. All p-values are uncorrected for multiple
comparisons.