0469
Motion estimation and correction with joint optimization for wave-CAIPI acquisition1Techna Institute, University Health Network, Toronto, ON, Canada, 2Koerner Scientist in MR Imaging, University Health Network, Toronto, ON, Canada, 3Center for Neuroscience Imaging Research, Institute for Basic Science & Department of Biomedical Engineering, Sungkyunkwan University, Suwon, Korea, Republic of
Synopsis
Wave-CAIPI is a recently introduced parallel imaging method with high reduction factor and low g-factor penalty, thus is less prone to motion for the patients who cannot hold steady for long time. This study revealed that wave-CAIPI is still sensitive to during-scan motion, and proposed a joint optimization method to estimate motion and mitigate the introduced artifacts in wave-CAIPI images.
Introduction
MRI scanning is sensitive to subject motion due to lengthy data acquisition, which is longer than most types of physiological movement cycles [1]. In order to reduce scanning time and the impact from motion, parallel imaging methods have been introduced and applied in clinical applications. Among these methods, the recently introduced wave-CAIPI [2] achieves significantly lower g-factor penalty to image signal-to-noise ratio (SNR) than the traditional SENSE and GRAPPA methods. Thus, it is capable to achieve a high acceleration factor and reduce the scan time of the whole-brain anatomical scan with a sub-millimeter resolution from ~5 to less than 1 minute [3]. Even though the scan time has been significantly shortened, significant motion-related image degradation using wave-CAIPI acquisition can persist, for example, due to short sudden movement, in particular in the readout direction. This study proposes and investigates the possibility of using joint optimization to estimate and correct motion for accelerated 3D wave-CAIPI acquisition.Methods
Data were acquired on a 3T MRI scanner (Prisma, Siemens AG, Erlangen, Germany) with a 32-channel head coil. A grapefruit was used as phantom, with the possibility of moving it from the outside of the magnet bore. The motion of the phantom was limited to translational movement within ±5 mm.Data acquisition: The GRE sequence with wave-encoding and butterfly navigator [4] features was in-house developed using IDEA. The common parameters for all scans were: TR 17ms, TE 8.5 ms, flip angle 20 degrees, matrix size 192×192×96 with 1 mm isotropic voxel size. For all the wave-CAIPI sequence, a sin and a cos wave were applied on Y and Z gradient axis, respectively, during the readout window with 7 cycles and 6 mT/m in amplitude. The following sequences were implemented in this study: (1) Full sampled Cartesian GRE without motion, scan time 5 min 15 s; (2) Accelerated wave-encoded GRE (Ry = 2, Rz = 4, kz blip = 2) without motion, scan time ~ 40 s; (3) Accelerated wave-CAIPI GRE (Ry = 2, Rz = 4, kz blip = 2) with motion, scan time ~ 40s. Butterfly navigators were acquired for sequence 3.
Data processing: Image reconstruction and motion estimation/correction were performed in MATLAB (R2018b, MathWorks, MA, USA). Coil sensitivity map was calculated with Berkeley Advanced Reconstruction Toolbox (BART) through ESPIRiT [5], and the SENSE-type reconstruction was used for the wave-CAIPI datasets.
The forward model $$$E$$$ that transforms image to each k-space line with consideration of rigid-body motion $$$\phi$$$ and wave-encoding phase $$$P$$$ is:
$$s=Ex=UF_{p}CT_{\phi}R_{\phi}x$$
where $$$x$$$ is the original object, $$$s$$$ is the acquired k-space data, $$$U$$$ is the k-space location mask, $$$F$$$ is Fourier transform operator with extra wave-encoding phase $$$P$$$ along the readout direction, $$$C$$$ is coil sensitivity map, $$$T$$$ and $$$R$$$ are translation and rotation operators due to motion trajectory $$$\phi$$$. The initial values of motion were determined by the butterfly navigator instead of random assigning for faster convergence in optimization.
Joint optimization with the following target function was used to the selected target voxels for simultaneously estimating the image $$$x$$$, wave-encoding phase $$$P$$$, and motion trajectory $$$\phi$$$:
$$argmin_{x,P,\phi}||s-Ex||_{2}^{2}$$
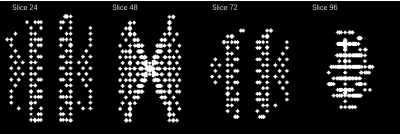
The point spread image of a single voxel was simulated, given the same under-sampling pattern, theoretical wave-encoding as sequence 3, and the motion initial values. This point spread image was used for selecting the pattern of target voxels in 3D image domain, and this selected pattern was moved in each iteration during optimization to cover the whole spatial area, similar as in ref [6].
Results and Discussion
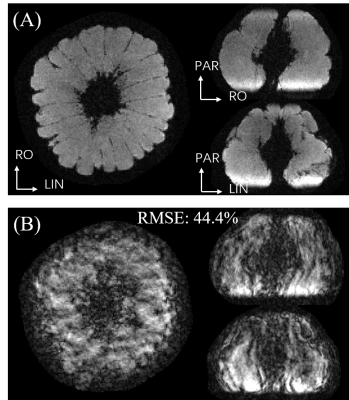
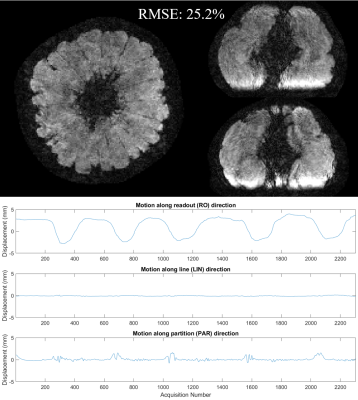
Figure 1 shows the comparison of the direct reconstruction results between no motion and motion-impacted wave-CAIPI 8× accelerated dataset. Unlike Cartesian images, motion artifact of wave-CAIPI images are not only along phase encoding directions, but also along readout direction. Figure 2 shows the point spread pattern under the scanning condition of sequence 3 on four slices. The point spread pattern is only along PE directions in Cartesian case. In contrast, in wave-CAIPI case, this point spread pattern is also along readout direction with a larger coverage, allowing smaller times of pattern shifting to cover the whole 3D FOV. Figure 3 shows the motion corrected wave-CAIPI images and the estimated motion trajectory. While mild level of motion artifacts still remain, image quality was dramatically improved, making the images comparable to the “no motion” ones. The root-mean-squared error to the non-motion wave-CAIPI scan reduced from 44.4% to 25.2%Conclusion
Parallel imaging using wave-CAIPI significantly reduced scanning time with low g-factor penalty to image SNR. However, it is still sensitive to within-scan motion and the motion artifacts are along all three spatial dimensions. This study revealed that the motion in wave-CAIPI images could be estimated and mitigated through joint optimization on targeted voxels selected by the simulation of point spread pattern. Accuracy and algorithm performance are currently optimized in an ongoing study.Acknowledgements
KU received funding from IBS (#IBS-R015-D1). The authors appreciate the support from Dr. Morgan Barense, Dr. Ali Golestani and Ms. Priya Abraham from Toronto Neuroimaging Facility (ToNI), Department of Psychology, University of Toronto.References
[1] Maxim Zaitsev, Julian Maclaren, and Michael Herbst. Motion Artifacts in MRI: A Complex Problem With Many Partial Solutions. Journal of Magnetic Resonance Imaging 42:887–901 (2015)
[2] Berkin Bilgic, Borjan A. Gagoski, Stephen F. Cauley, Audrey P. Fan, Jonathan R. Polimeni, P. Ellen Grant, Lawrence L. Wald, and Kawin Setsompop. Wave-CAIPI for Highly Accelerated 3D Imaging. Magnetic Resonance in Medicine 73:2152–2162 (2015)
[3] Daniel Polak, Kawin Setsompop, Stephen F. Cauley, Borjan A. Gagoski, Himanshu Bhat, Florian Maier, Peter Bachert, Lawrence L. Wald, and Berkin Bilgic. Wave-CAIPI for Highly Accelerated MP-RAGE Imaging. Magnetic Resonance in Medicine 79:401–406 (2018)
[4] Joseph Y. Cheng, Marcus T. Alley, Charles H. Cunningham, Shreyas S. Vasanawala, John M. Pauly, and Michael Lustig. Nonrigid Motion Correction in 3D Using Autofocusing With Localized Linear Translations. Magnetic Resonance in Medicine 68:1785–1797 (2012)
[5] Martin Uecker, Peng Lai, Mark J. Murphy, Patrick Virtue, Michael Elad, John M. Pauly, Shreyas S. Vasanawala, and Michael Lustig. ESPIRiT - An Eigenvalue Approach to Autocalibrating Parallel MRI: Where SENSE meets GRAPPA. Magnetic Resonance in Medicine, 71:990-1001 (2014)
[6] Melissa W. Haskell, Stephen F. Cauley, and Lawrence L. Wald. TArgeted Motion Estimation and Reduction (TAMER): Data Consistency Based Motion Mitigation for MRI Using a Reduced Model Joint Optimization. IEEE Transactions on Medical Imaging, 37(5): 1253-1264 (2018)
Figures