0450
Fully-automated 1H MRI Thoracic Cavity Segmentation for Hyperpolarized Gas Imaging using a Convolutional Neural Network1Medical Biophysics, Robarts Research Institute, Western University, London, ON, Canada, 2School of Biomedical Engineering, Robarts Research Institute, Western University, London, ON, Canada
Synopsis
Thoracic segmentations are crucial for accurate measurements of normalized lung ventilation, perfusion and gas exchange. Current semi-automated methods are time consuming, require experienced readers, and lack the standardization of fully-automated methods, such as convolutional neural networks. We retrospectively pooled data from 449 healthy and respiratory disease participants, resulting in a 55,000 slice augmented data set to train a dense v-net neural network. The network produced segmentations qualitatively matching semi-automated methods, with high Dice scores and an area under the receiver operating characteristic curve of 0.997. Implementation on the NiftyNet platform permits quick model dissemination for multi-site validation.
PURPOSE
Hyperpolarized gas MRI (3He and 129Xe) has allowed advancements in the understanding of lung pathophysiology and mechanics, especially through the ventilation defect percent (VDP) biomarker, defined as the ratio of defect volume to lung volume. Volume-matched 1H MRI are acquired alongside hyperpolarized MRI to segment the thoracic cavity for VDP. Prior methods for thoracic segmentation have relied on semi-automated methods of user seeding and correction;1,2 a truly automatic method is desirable for repeatability, multi-site studies, and clinical deployment. Recently, convolutional neural networks (CNN) have shown promise in providing precise, fast and fully automated segmentations in medical imaging. Despite these advances, we are not aware of previously published CNN methods for MRI lung segmentation. We hypothesized that a dense v-net would be able to produce significantly accurate segmentations of the thoracic cavity. The dense v-net architecture is optimized for multi-class thoracic segmentations,3 making it ideal for identification of right and left lungs. The objective of this work was to train a dense v-net CNN and validate network segmentations using Dice similarity coefficients (DSC) and receiver operating characteristics (ROCs).Methods
Participants and Image Acquisition:Data were retrospectively pooled from previous imaging studies. 1H thoracic MRI were acquired using a 3.0T Discovery MR750 system (GEHC, USA). Participants were placed supine and inhaled 1.0L of N2. Coronal whole lung images were acquired using a FGRE sequence (acquisition time: 16s; TR/TE/flip=3.7ms/0.956ms/20o; FOV=40x40cm2; matrix=128x128; slice thickness=15mm).
Data Annotation:
The data compilation process is shown in Figure 1. Data were pooled from previous hyperpolarized imaging studies in healthy participants, ex-smoker participants with and without chronic obstructive pulmonary disease (COPD), and participants with asthma. This was beneficial for training the network across multiple health states. 3D convolutional networks perform poorly on anisotropic voxels, therefore the volumes were divided into 15-17 2D images yielding 27612 data sets. Axis-swapping augmented the data to 55224 sets. Ground-truth labeling was performed by 4 trained imaging scientists with 0.5-4 years (mean=2yrs) experience in ventilation imaging using a semi-automated segmentation tool1 that provided an initial segmentation estimate using an automated region growing algorithm followed by manual correction.
Network:
A dense v-net architecture was adapted from the NiftyNet platform,4 based on TensorFlow libraries, and implemented on a workstation GPU (NVidia 11GB GTX1080Ti). Whole-images were used for random sampling (window=128x128x1, queue=160, batch=32 per iteration). Training took place over 10000 iterations (equivalent to 8 epochs). Network learning was driven by a combination Dice and cross-entropy loss function.5 Data were split into development and inference groups. The model was validated using 5-fold cross validation of the development group (ntraining=314, nvalidation=45, ntesting=90). Output segmentations were evaluated to calculate sensitivity, specificity, dice coefficient and area under the curve (AUC) for ROC.
Results
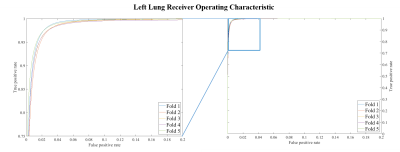
Model testing occurred in approximately 0.08s/volume when executed via GPU. Figure 2 shows example segmentations from the 1st-fold testing step. There was good qualitative agreement between ground-truth and CNN results. CNN segmentations tended to slightly over-estimate lung volume, bleed into the diaphragm on occasion, and were relatively insensitive to pulmonary arteries. Quantitative analyses showed exceptional segmentation overlap (average DSC=0.96, DSC=0.95 for right and left lungs). Segmentation effectiveness was slice dependent (DSCslice-1=0.73 for both, DSCslice-15 =0.46, 0.66 respectively) with central slices in better agreement with ground truth (DSCslice-8=0.97 for both). ROC in figure 3 demonstrate that the model was highly sensitive and specific (average AUC=0.997).Discussion
CNN performance allowed fully-automated left and right lung segmentation in a fraction of the time required for current semi-automated methods. Data were skewed towards diseased states, however this matches the intended application of the network. Model results experienced occasional imperfections around pulmonary arteries: adding manual labels for arteries during training may improve performance in future. Model performance was most poor at the most anterior and posterior slices (slices 1-2 and 13-15) due to a combination of partial volume effects and difficulty when the anterior mediastinum was thin – readers labeled this as a continuous region. The impact of reduced anterior and posterior performance is minimal since current analyses tend to focus on center-slice defects. Model uncertainty is greatest in partial-volume regions. By adding hyperpolarized gas images in future, information about the presence or absence of gas may improve partial-volume segmentation.Conclusions
A dense v-net was implemented that successfully and accurately segmented left and right lungs in proton MRI. Quantitative DSC and ROC analyses indicated results matched semi-automated ground truths. The NiftyNet platform allows simple distribution of a trained model and dependencies,4 which may permit multi-site implementation in future.Acknowledgements
No acknowledgement found.References
1. Kirby, M. et al. Acad Radiol 19, 141-152 (2012).
2. Guo, F. et al. Med Image Anal 23, 43-55 (2015).
3. Gibson, E. et al. IEEE Trans Med Imaging 37, 1822-1834 (2018).
4. Gibson, E. et al. Comput Methods Programs Biomed 158, 113-122 (2018).
5. Isensee, F. et al. nnU-Net: Self-adapting Framework for U-Net-Based Medical Image Segmentation. arXiv e-prints (2018). <https://ui.adsabs.harvard.edu/abs/2018arXiv180910486I>.
Figures