0426
MRI-based Radiomics as a Predictive Biomarker of Survival in High Grade Gliomas Treated with Chimeric Antigen Receptor T-Cell Therapy
Sohaib Naim1, Chi Wah Wong2, Eemon Tizpa1, Hannah Jade Young1, Kimberly Jane Bonjoc1, Seth Michael Hilliard1, Aleksandr Filippov 1, Saman Tabassum Khan1, Christine Brown3, Behnam Badie4, and Ammar Ahmed Chaudhry1
1Diagnostic Radiology, City of Hope National Medical Center, Duarte, CA, United States, 2Applied AI and Data Science, City of Hope National Medical Center, Duarte, CA, United States, 3Hematology & Hematopoietic Cell Transplantation and Immuno-Oncology, City of Hope National Medical Center, Duarte, CA, United States, 4Surgery, City of Hope National Medical Center, Duarte, CA, United States
1Diagnostic Radiology, City of Hope National Medical Center, Duarte, CA, United States, 2Applied AI and Data Science, City of Hope National Medical Center, Duarte, CA, United States, 3Hematology & Hematopoietic Cell Transplantation and Immuno-Oncology, City of Hope National Medical Center, Duarte, CA, United States, 4Surgery, City of Hope National Medical Center, Duarte, CA, United States
Synopsis
High grade gliomas (HGG) is the most common malignant primary brain tumors in adults. In this study, 61 patients with recurrent HGGs underwent surgical resection and chimeric antigen receptor-T cell therapy. Volumetric segmentations of contrast-enhanced (CE) and non-enhanced tumors (NET) using T1-weighted CE MR images were used to identify shape- and texture-based features from these regions of interest. We evaluated radiomic characteristics of these HGGs to determine novel imaging biomarkers to predict treatment response. Exponentially-filtered textural radiomic features based on Neighboring Gray Tone Difference Matrix and Gray Level Co-occurrence Matrix derived from NET were the strongest predictors of overall survival.
Introduction
High grade glioma (HGG) is the most common primary malignant brain tumor that is rapidly progressive and fatal. In the era of targeted immunotherapy in neuro-oncology, there is rapid emergence of novel molecular and cellular therapies (e.g. pembrolizumab, Chimeric Antigen Receptor [CAR] T-cell therapy). There is an immediate unmet need of noninvasive biomarkers predictive of treatment response. In this study, we evaluate the MRI-based radiomic characteristics of HGG treated with CAR T-cell therapy with aim to identify novel shape- and texture-based features predictive of positive treatment response.Materials and Methods
In this prospective IRB approved phase I study, 61 patients suffering from HGG underwent surgical resection and CAR T-cell therapy.1 All patients underwent baseline MRI scans prior to surgical resection and CAR T-cell administration in the resection cavity. The workflow of the radiomic analysis is summarized in Figure 1. In summary, we utilized Brain Tumor Image Analysis (BraTumIA v. 2.0.0.5) software generating automatic volume segmentations of healthy and tumor tissue incorporating T1- and T2-weighted sequences. BraTumIA generated labels were then manually segmented for contrast enhanced (CE) tumors and non-enhanced tumors (NET). Our model focuses on identifying shape-based, texture-based and image-filtered radiomic features that were extracted from the CE and NET regions of interest (ROI) using T1-weighted images.2 Logistic regression was used to classify whether survival is above or below 200 days, which is the median survival. To avoid over-fitting, elastic-net regularization and leave-one-out cross-validation were used.Results
Using the CE and NET tumor ROIs, we identified 2453 radiomic features for predictive modeling. Our classification model’s performance predicting survival is shown in Figure 2, which shows a validation Area Under the Receiver Operating Characteristic Curve (AUC) of 0.74. The top 5 predictive radiomic biomarkers are texture-based derived from exponentially filtered non-enhancing tumor segmentations and are displayed in Figure 3.Discussion and Conclusion
Our radiomic feature extraction evaluating CAR T-cell treated HGG identified neighboring gray tone difference matrix texture-based feature as the strongest predictor of overall survival (OS). Novel and effective biomarkers evaluated through radiomic feature-analysis can provide a more personalized response assessment for patients suffering from HGG. In future work, we can incorporate T2-based features along with clinical and molecular information that may better predict survival.Keywords
High grade glioma (HGG), CAR-T therapy, Imaging Biomarkers, Tumor Segmentation, RadiomicsAcknowledgements
No acknowledgement found.References
- Brown CE, Alizadeh D, Starr R, Weng L, Wagner JR, Naranjo A, Ostberg JR, Blanchard MS, Kilpatrick J, Simpson J, Kurien A, Priceman SJ, Wang X, Harshbarger TL, D'Apuzzo M, Ressler JA, Jensen MC, Barish ME, Chen M, Portnow J, Forman SJ, Badie B. Regression of Glioblastoma after Chimeric Antigen Receptor T-Cell Therapy. N Engl J Med. 2016 Dec 29;375(26):2561-9. doi: 10.1056/NEJMoa1610497.
- Pyradiomics Package (van Griethuysen, J. J. M., Fedorov, A., Parmar, C., Hosny, A., Aucoin, N., Narayan, V., Beets-Tan, R. G. H., Fillon-Robin, J. C., Pieper, S., Aerts, H. J. W. L. (2017). Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Research, 77(21), e104–e107. `https://doi.org/10.1158/0008-5472.CAN-17-0339 <https://doi.org/10.1158/0008-5472.CAN-17-0339>`_)
Figures
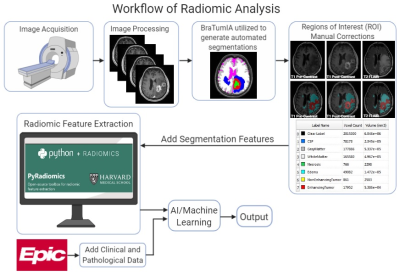
The acquired images from the scanners are automatically segmented
using BraTumIA (BraTumIA v. 2.0.0.5) and manually corrected using ITK-SNAP. Radiomic
features are extracted using PyRadiomics.2 All features are merged
together for a machine learning model to predict outcome. In this study, we
only considered contrast-enhanced (CE) and non-enhanced tumors (NET) using
T1-weighted images.
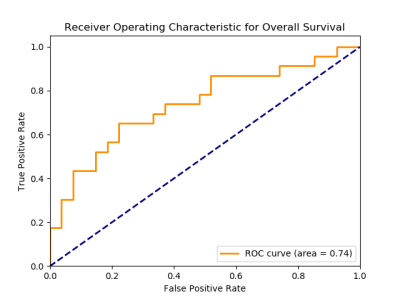
The ROC curve illustrates the classification model
performance to predict overall survival in HGGs treated with CAR-T cell
therapy.
Top 5 Radiomic features that are the most predictive of
overall survival in HGGs treated with CART cell therapy. Features derived from
the Gray level co-occurrence matrix (GLCM) and neighboring gray tone difference
matrix (NGTDM) were found to be the most predictive.2