0423
Using anatomic and diffusion MRI with deep convolutional neural networks to distinguish treatment-induced injury from recurrent glioblastoma1Radiology and Biomedical Imaging, UCSF, San Francisco, CA, United States, 2Bioengineering and Therapeutic Sciences, UCSF, San Francisco, CA, United States, 3UCSF, Neurological Surgery, CA, United States, 4Data Science, USF, San Francisco, CA, United States, 5Neurological Surgery, UCSF, San Francisco, CA, United States, 6Graduate Program in Bioengineering, UCSF/UC Berkeley, San Francisco and Berkeley, CA, United States
Synopsis
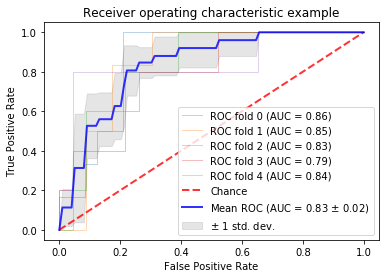
In this study, we leverage a promising new centrally restricted diffusion pattern1 together with modern advances in deep learning to create a novel method for detecting treatment-related injury in the context of suspected recurrent glioblastoma. We report a 5-fold cross-validation average AUC ROC of 0.83 +/- 0.2 for the classification of lesions into two categories: those induced by treatment, and those that are true incidences of recurrent glioblastoma.
Introduction
It is estimated that 25% to 35% of patients with high-grade glioma experience treatment-associated injury that can mimic recurrence[2],[3], posing a significant diagnostic challenge for radiologists. It is critical to distinguish these phenomena in order to accurately assess patient response to therapy. The presence of centrally restricted diffusion in the necrotic region of potentially recurrent high-grade gliomas has recently shown promise in assessing the presence of treatment-associated injury[1]. The goal of this study was to assess whether exploitation of this pattern in combination with recent advances in artificial intelligence will provide faster classification and enhanced sensitivity and specificity compared with visual assessment of the presence of this phenomenon.Methods
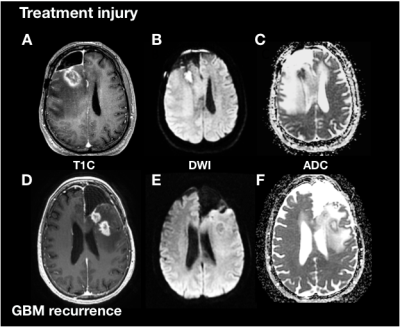
Subjects and Image Acquisition: 144 patients with suspected recurrent glioblastoma were scanned on a 3T scanner with an 8-channel head coil. T2-weighted FLAIR (T2 FLAIR) and 3D T1-weighted IR-SPGR imaging before (T1) and after(T1C) the injection of a gadolinium-based contrast agent were acquired. Diffusion-tensor images (DTI) were obtained in the axial plane with either 6 directions and 4 excitations or ≥24 directions and 1 excitation [TR/TE = 1000/108 ms, voxel size = 1.7 × 1.7 × 3 mm, b=1000 or 2000 s/mm] and apparent diffusion coefficient (ADC) maps were generated using FMRIB’s Diffusion Toolkit[4].Immunohistochemistry: A board-certified pathologist evaluated multiple tissue samples per patient and scored them on the presence of tumor cells. Tumors containing exclusively tissue with no viable tumor and signs of treatment effect (e.g. gliosis, hyalinized blood vessels) were given the designation of “treatment effect” (TxE), while patients with recurrent tumor were diagnosed at the time of surgery with > 25% viable tumor. With this approach, 27 patients were classified as having treatment effect, and 117 patients were classified as having recurrent glioblastoma.
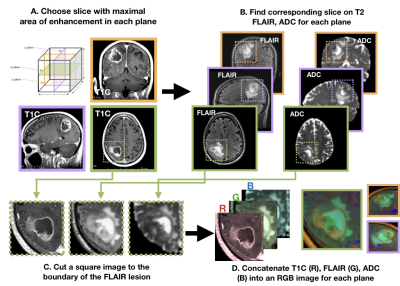
Image processing: Each image was multiplied by a brain mask and z-score normalization was performed. The maximum area of the contrast enhancing lesion (CEL) or nonenhancing lesion (NEL) as determined by the ROI area was used to select the slice of interest in each plane (Figure 1). A square box padding the bounds of the NEL was used to extract a patch from the T1C, T2 FLAIR, and ADC images. These patches from each MR modality were concatenated to generate a single RGB-color image for each plane (coronal, sagittal and axial), for each patient.
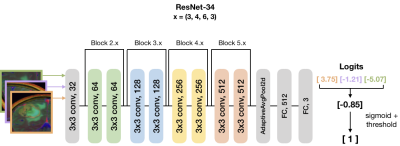
Training, testing, model and computational framework: Patients were split into 5 folds and stratified based on outcome. A ResNet-34 pre-trained with ImageNet was used to initialize the weights for model training (Figure 2)[5]. During training, the minority (TxE) class was oversampled 4x through data augmentation to account for the fewer number of patients in the TxE group. Data was augmented using cropping, rotating, and flipping both horizontally and vertically. Cosine differential learning rates (LR) and cross-entropy loss functions were used during training. The logit output of the final network layer was averaged across the three directions for each patient and a sigmoid function was applied. All model building, training, and testing were implemented using PyTorch 1.0.0 on a Tesla V100-PCIE-32GB GPU (Nvidia).
Results + Discussion
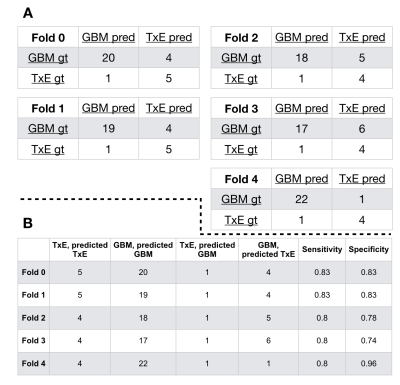
Our pre-trained ResNet-34 model resulted in a 5-fold cross-validation average AUC ROC of 0.83 +/- 0.2 for the classification of patients into their respective categories (Figure 4.) Figure 5 describes the confusion matrices achieved when choosing a threshold corresponding to the top left point on the AUC ROC curve. Our average sensitivity and specificity, 81.3% and 82.7%, are in line with 75% and 89% reported by Zakhari et al.[1]; however, our cohort includes over 8 times as many patients, and 3 times the number of TxE lesions. The vast majority of prior efforts in this field use radiomics or calculate the median diffusion-, perfusion-, or spectroscopic- derived parameter value from an anatomical ROI to obtain a threshold that can distinguish treatment effects from true tumor recurrence [6,7]; however, these methods suffer from either requiring manual intervention for selecting regions of interest, lack of pathological confirmation, low sample size, or assessing associations rather than prediction ability. Our approach is unique in that it leverages: 1) multiple tissue samples for generating a single pathological outcome in the analysis, 2) transfer learning with the merging multi-contrast images into one input, 3) a substantially larger dataset than prior studies, and 4) a fully automated pipeline.Conclusion
In this study, we leverage a promising new centrally restricted diffusion pattern[1] together with modern advances in deep learning to create a novel strategy for detecting treatment-associated injury in the context of suspected recurrent glioblastoma. Through combining clinical diffusion-weighted imaging with standard anatomical imaging, it was possible to build an algorithm that generated imaging features relevant in distinguishing true recurrent glioblastoma from treatment-associated injury. Data collection of an independent test is currently underway to test the generalizability of our trained network on unseen data.Acknowledgements
No acknowledgement found.References
1. Zakhari N, Taccone MS, Torres C, et al. Diagnostic Accuracy of Centrally Restricted Diffusion in the Differentiation of Treatment-Related Necrosis from Tumor Recurrence in High-Grade Gliomas. AJNR Am J Neuroradiol 2018;39(2):260–4.
2. Ellingson BM, Wen PY, Cloughesy TF. Modified criteria for radiographic response assessment in glioblastoma clinical trials. Neurotherapeutics 2017;14(2):307–20.
3. Abbasi AW, Westerlaan HE, Holtman GA, Aden KM, van Laar PJ, van der Hoorn A. Incidence of Tumour Progression and Pseudoprogression in High-Grade Gliomas: a Systematic Review and Meta-Analysis. Clin Neuroradiol 2017;28(3):401–11.
4. Duarte-Carvajalino JM, Sapiro G, Harel N, Lenglet C. A Framework for Linear and Non-Linear Registration of Diffusion-Weighted MRIs Using Angular Interpolation. Front Neurosci 2013;7:41.
5. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Proceedings of 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). IEEE; 2016. p. 770–8.
6. van Dijken BRJ, van Laar PJ, Holtman GA, van der Hoorn A. Diagnostic accuracy of magnetic resonance imaging techniques for treatment response evaluation in patients with high-grade glioma, a systematic review and meta-analysis. Eur Radiol 2017;27(10):4129–44.
7. Verma N, Cowperthwaite MC, Burnett MG, Markey MK. Differentiating tumor recurrence from treatment necrosis: a review of neuro-oncologic imaging strategies. Neuro Oncol 2013;15(5):515–34.
Figures