0422
Identifying Overall Survival in Glioblastoma Patients Using VASARI Features at 3T1Institute of Biomedical Engineering, Bogazici University, Istanbul, Turkey, 2Department of Radiology, Yeditepe University, Istanbul, Turkey, 3Department of Neurosurgery, Yeditepe University, Istanbul, Turkey
Synopsis
Glioblastoma (GBM) is the most common primary brain tumor in adults with 15 months median overall survival. The purpose of this study was to identify overall survival of GBM patients based on clinical and Visually AcceSAble Rembrandt Images (VASARI) features using machine learning. According to our results, a support vector machine (SVM) model worked better for categorical data classification. With the help of adaptive synthetic (ADASYN) oversampling, a fine Gaussian SVM model identified short overall survival at 12 and 24 months thresholds with 99.78% and 88.80% accuracies, respectively.
Introduction
Glioblastoma (GBM), constituting 80% of primary brain tumors, is the most malign central nervous system tumor1. Median overall survival from GBM is less than 15 months2 despite all medical interventions. The most important factor in improving the quality of life for GBM patients is an effective treatment plan, which highly depends on estimating their survival. Recently, Visually Accessible Rembrandt Images (VASARI) analysis has been developed to provide a qualitative assessment of MR images of gliomas3. This imaging feature system comprises 26 features each represented by a feature name like tumor location, proportion of necrosis, and T1/FLAIR ratio, and a corresponding score number. In this study, VASARI features and some clinical data of GBM patients were used in training several machine learning algorithms to predict survival.Methods
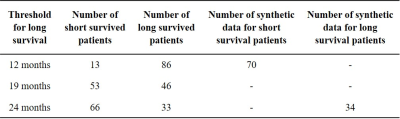
Ninety-nine GBM patients (67M/32F, mean age: 53.95±13.81 years, range: 11-88 years) were included in this retrospective study. The patients were scanned at a 3T Philips Ingenia scanner (Best, Netherlands) using a standard brain tumor MRI protocol that included a T1-weighted 3D turbo field echo (TFE) sequence (TR=9.1ms, TE=4.2ms, flip angle=8, slice thickness=1 mm, gap=0.3 mm), T2-weighted (TR=11000ms, TE=125ms, TI=2800ms, slice thickness=4 mm, gap=1 mm), and diffusion-weighted MRI (TR=4574ms, TE=55ms, EPI factor=47, slice thickness=4 mm, gap=1 mm, slice number=28, scan time=59 s, b=1000 s/mm2)., diffusion tensor imaging (TR=10,000ms, TE=53ms, EPI factor=67, slice thickness=2.5 mm, gap=0 mm, slice number=60, scan time=6 min), and perfusion MRI (TR=1500ms, TE=50ms, flip angle=40°, slice thickness=4 mm, slice gap=1 mm, 45 dynamics, scan time=1.2 min). MRI of each patient were scored by an experienced neuroradiologist using VASARI feature system as previously described3. The dataset used in this study was comprised of some clinic features (age, gender, extent of resection, pre- and post-KPS, ki67 and P53) and 26 VASARI imaging features. The patients were classified into two groups, as with low or high survival. The labeling of the dataset was performed considering three different survival threshold values (12, 19, and 24 months). The dataset was imbalanced for the thresholds of 12 and 24 months. So, adaptive synthetic (ADASYN)4 oversampling method, which generates synthetic data from the real data based on k-nearest neighbors algorithm, was used for handling these imbalanced datasets. Machine learning models defined in Classification Learner app in MATLAB 2019a (The MathWorks Inc., Natick, MA) were employed to predict survival of GBM patients based on clinical and VASARI features. The survival prediction was performed for all the three survival thresholds. In all the classifications, the data was divided into training and testing sets with 10-fold cross validation. All the models were executed 100 times and the mean performance metrics were reported.Results
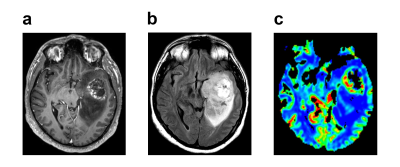
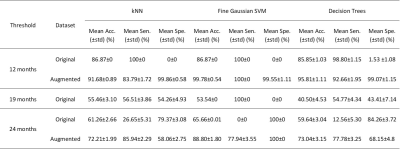
Figure 1 shows post-contrast T1-weighted (a), T2-weighted (b) and perfusion (c) MR images of a patient who had a high grade lesion at the left mesial temporal lobe with hemorrhagic component inside and surrounded by expansile edema. Table 1 shows the number of short and long survived patients along with the number of synthetic data at 12, 19, and 24 months thresholds. For 19 months threshold, there was no need to synthesize data, since the numbers of short and long survived patients were almost equal. For 12 and 24 months thresholds, 70 short survival and 34 long survival synthetic data points were produced with ADASYN, respectively. Table 2 summarizes the classification results of the machine learning models giving the highest accuracies for 3 different thresholds and both original and augmented data. For the 12 months threshold, the classification results were improved by oversampling and the best performance results were obtained with a fine Gaussian support vector machine (SVM) model resulting in an accuracy of 99.78%, sensitivity of 100%, and specificity of 99.55%. Similarly, for the 24 months threshold, a fine Gaussian SVM model resulted in the best classification performance, with an accuracy of 88.80%, sensitivity of 77.94%, and specificity of 100%. On the other hand, the machine learning models were not able to accurately classify the data labeled with a 19 months threshold.Discussion and Conclusion
The results of this study indicated the VASARI features along with the clinical data might be used to identify short overall survival in GBM. The data imbalance resulted in the classification of all the patients into the majority class, resulting in either high specificity (24 months) or sensitivity (12 months) in the original data. Oversampling enabled learning also from the minority classes, and improved the performance of the machine learning models in both datasets. SVM resulted in the best classification performance for all datasets, since the data was categorical. Future studies will be conducted on a larger patient cohort to determine if the survival of GBM patients could be identified better even without oversampling.Acknowledgements
No acknowledgement found.References
1. Agnihotri S, Burrell KE, Wolf A, et al. Glioblastoma, a Brief Review of History, Molecular Genetics, Animal Models and Novel Therapeutic Strategies. Archivum Immunologiae et Therapiae Experimentalis. 2013; 61: 25-41.
2. Thakkar JP, Dolecek TA, Horbinski C, et al. Epidemiologic and molecular prognostic review of glioblastoma. Cancer Epidemiol Biomarkers Prev. 2014; 23: 1985-1996.
3. Cancer Imaging Archive. VASARI Research Project. 2015.
4. Haibo H, Yang B, Garcia EA, Shutao L. ADASYN: Adaptive synthetic sampling approach for imbalanced learning. 2008 IEEE International Joint Conference on Neural Networks (IEEE World Congress on Computational Intelligence), 2008; 1322-1328.
Figures