0419
Fast multimodal image fusion with deep 3D convolutional networks for neurosurgical guidance – A preliminary study1General Electric Research, Niskayuna, NY, United States, 2Brigham and Women's Hospital, Boston, MA, United States
Synopsis
Multimodality fusion in neurosurgical guidance aids neurosurgeons in making critical clinical decisions regarding safe maximal resection of tumors. It is challenging to have registration methods that automatically update pre-surgical MRI on intra-operative ultrasound, adjusting for the brain-shift for surgical guidance. A 3D deep learning-based convolutional network was developed for fast, multimodal alignment of pre-surgical MRI and intra-operative ultrasound volumes. The neural network is a combination of some well-known deep-learning architectures like FlowNet, Spatial Transformer Networks and UNet to achieve fast alignment of multimodal images. The CuRIOUS 2018 challenge training data was used to evaluate the accuracy of the developed method.
INTRODUCTION
Multimodal image fusion in image guided neurosurgery aids neurosurgeons in making critical clinical decisions regarding safe maximal resection of tumors. The brain however undergoes substantial non-linear structural deformation on account of multiple factors, including dura opening and tumor resection, referred to as brain-shift. It is crucial and challenging to have methods that automatically register preoperative MRI (pre-MRI) to intra-operative ultrasound (iUS) to compensate for brain-shift during surgical guidance. These methods need to be fast (~3fps) and accurate (~2mm). In this work, we present a fast, deep learning (D/L)-based unsupervised deformable registration method that aligns pre-MRI on iUS for neurosurgical guidance.METHOD
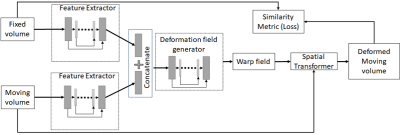
A new unsupervised 3D D/L-based method was developed to non-rigidly register pre-MRI to iUS brain images available from the CuRIOUS 2018 challenge training dataset1. The network architecture was based on UNet, FlowNet and Spatial Transformer Networks2-4 (Fig. 1). The novelty of the architecture is in the use of U-Net for both feature extraction and flow estimation as lower-level features are incorporated in the feature representation at higher-levels.In the training phase, the rigidly registered pre-MRI and iUS were passed through U-Net-based feature extractors and the features were then stacked and passed through another U-Net based optical flow estimator that estimated the deformation field between the pre-MRI and iUS. The pre-MRI was warped with the deformation field, and the normalized cross correlation between the warped pre-procedure MRI and iUS was maximized as part of optimization via back propagation. In the prediction phase, given a paired pre-MRI and iUS, deformation fields were predicted, and the pre-MRI was warped in less than 3 secs on a NVIDIA V100 GPU with 32GB memory.
A total of 22 datasets are available in CuRIOUS 2018 training data. Each data set has a gadolinium-enhanced T1-weighted (T1w) and a T2-weighted-FLAIR (T2w) MRI acquired preoperatively, and an intraoperative 3D iUS image acquired after dura opening. The T2w-FLAIR and iUS images were used for the demonstration of fast multi-modal registration.
Data from 18 of the 22 subjects were randomly chosen for training the neural network and the prediction of the deformation field was validated on the remaining 4 subjects. The process was performed twice in a cross-validation framework resulting in the prediction of deformation field in 8 subjects. 14 homologous landmarks available for each dataset of the challenge data were used to evaluate the accuracy of the deformable registration.
RESULTS
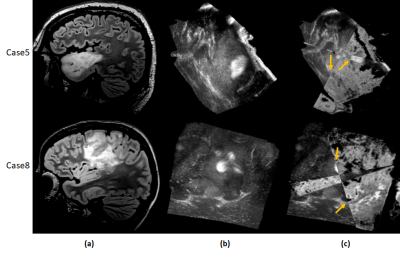
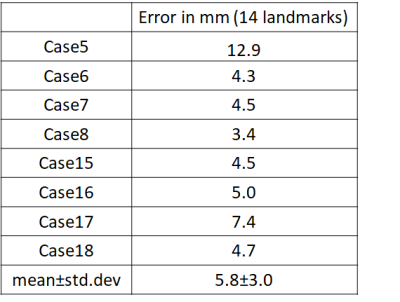
Table 1 shows the mean error in aligning the T2w-FLAIR to the iUS for 14 landmarks in each of the 8 datasets. A mean error of 5.8mm ± 3.0mm was achieved for all landmarks and all 8 validation cases in 2.6 secs per case. All methods in the CuRIOUS challenge achieved errors in the range of 1.5mm - 6.5mm with registration time in the range of 1.8 secs - 450 secs5. Fig.2 shows the quality of alignment for two subjects Case5 and Case8 respectively of the CuRIOUS 2018 challenge.DISCUSSIONS AND CONCLUSIONS
This is a preliminary study on the accuracy of a D/L-based network that is fast in predicting alignment between pre-MR and iUS. D/L based methods are fast compared to traditional registration methods and therefore are ideal for image-fusion guided interventions; however, these methods perform well when trained with large number of datasets. We believe with enough data we will be able to optimize the network and achieve target registration accuracies of less than 2mm.*Soumya Ghose and Jhimli Mitra are joint first authors on this abstract.
Acknowledgements
No acknowledgement found.References
1) Xiao Y, Rivaz H, Chabanas M, Fortin M, Machado I, Ou Y, Heinrich MP, Schnabel JA, Zhong X, Maier A, Wein W, Shams R, Kadoury S, Drobny D, Modat M, Reinertsen I, Evaluation of MRI to ultrasound registration methods for brain shift correction: The CuRIOUS2018 Challenge, arXiv:1904.10535.
2) Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Proc of the Medical Image Computing and Computer-Assisted Intervention (MICCAI), Springer, 2015, LNCS, vol.9351: 23-241.
3) Dosovitskiy A, Fischer P, Ilg E, Häusser P, Hazırbaş C, Golkov V, FlowNet: learning optical flow with convolutional networks. In:Proc of the IEEE International Conference on Computer Vision, 2015, pp. 2758–2766.
4) Jadenberg M, Simonyan K, Zisserman A, Kavukcuoglu K, Spatial Transformer Networks. In: Proc of the 28th Intl Conf. on Neural Information Processing Systems -vol2, 2015, pp. 2017-2025.
5) Xiao Y, Rivaz H, Chabanas M, Fortin M, Machado I, Ou Y, Heinrich MP, Schnabel JA, Zhong X, Maier A, Wein W, Shams R, Kadoury S, Drobny D, Modat M, Reinertsen I, Evaluation of MRI to ultrasound registration methods for brain shift correction: The CuRIOUS2018 Challenge. IEEE Transactions on Medical Imaging, Epub, 2019.
Figures