0409
Statistical multiscale mapping of IDH1, MGMT, and microvascularity in human brain tumors from multiparametric MR and registered core biopsy1Radiology & Imaging Sciences, Indiana University School of Medicine, Indianapolis, IN, United States, 2School of Health Sciences, Purdue University, West Lafayette, IN, United States, 3Biostatistics, Indiana University School of Medicine, Indianapolis, IN, United States, 4Military Health Institute, University of Texas Health San Antonio, San Antonio, TX, United States, 5Neuroradiology, Lucile Salter Packard Children’s Hospital and Stanford University Medical Center, Palo Alto, CA, United States, 6Neurosurgery, Dayton Children's Hospital, Dayton, OH, United States
Synopsis
We demonstrate statistical relationships between routine multiparametric imaging signatures and underlying cellular and molecular properties of brain tumors. We apply advanced statistical methods to correct for the family-wise error rate problem associated with whole-brain statistical parametric mapping, and show that the results have strong agreement with surgical biopsy. These results imply that cellular and molecular mapping of tumor heterogeneity from minimally-invasive images may be possible in the near future.
INTRODUCTION
Emerging targeted therapies interfere with specific molecules that promote tumor growth and infiltration based on patient-specific predictive cellular and molecular biomarkers [1]. However, heterogeneous genomic and phenotypic tumor microenvironments contribute to incomplete treatment by targeted therapy and promote tumor recurrence via a non-linear branched evolution of the cancer genome [2]-[4]. Biopsy is currently the most effective method to assess patient-specific tumor biomarkers for targeted therapeutics, but clinical outcomes are limited by tumor heterogeneity which cannot be assessed by invasive biopsy alone [1]. Medical imaging techniques that are minimally-invasive and assess cellular and molecular tissue characteristics across the entire tumor bed and tumor microenvironment (TME) hold the potential to significantly improve the characterization and treatment of aggressive brain tumors [5]-[7]. Here we propose to map cellular and molecular tumor properties throughout the TME in a voxel-wise manner by leveraging the growing dimensionality of clinical MR data. Our approach does not inherently rely on spatial correlation information or simulations of various tissue properties for classification.METHODS
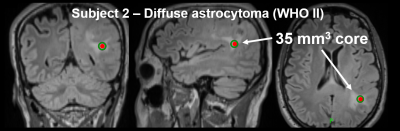
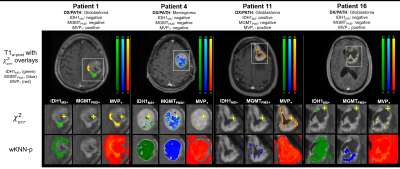
Twenty-nine patients underwent pre-surgical mp-MR followed by MR-guided stereotactic core biopsy. All mp-MR acquisitions included T1w, T2w, T2w-FLAIR, DWI, and T1w-post-gad. The locations of the biopsy cores were identified in the pre-surgical images using stereotactic bitmaps acquired during surgery. Feature matrices mapped the multiparametric voxel values in the vicinity of the biopsy cores to the pathologic outcome variables for each patient and logistic regression tested the individual and collective predictive power of the MR contrasts. A non-parametric weighted k-nearest neighbor classifier evaluated the feature matrices in a leave-one-out cross validation design across patients. Resulting class membership probabilities were converted to chi-square statistics to develop full-brain parametric maps, implementing Gaussian random field theory to estimate inter-voxel dependencies. Corrections for family-wise error rates were performed using Benjamini-Hochberg and random field theory, and the resulting accuracies were compared.RESULTS
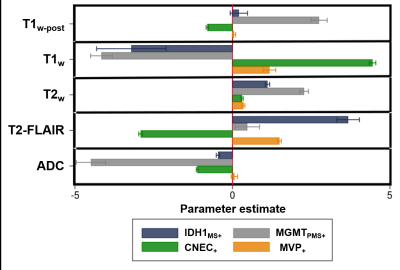
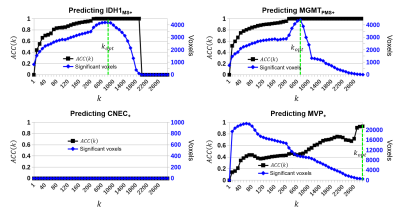
The combination of all five image contrasts correlated with outcome (P<10-4) for all four microscopic variables. The probabilistic mapping method using Benjamini-Hochberg generated statistically significant results (α<.05) for three of the four dependent variables: 1) IDH1, 2) MGMT, and 3) microvascular proliferation, with an average classification accuracy of 0.984 ± 0.02 and an average classification sensitivity of 1.567% ± 0.967. The images corrected by random field theory demonstrated improved classification accuracy (0.989 ± 0.008) and classification sensitivity (5.967% ± 2.857) compared with Benjamini-Hochberg.DISCUSSION
Cellular and molecular heterogeneity is a significant driver of brain tumor morbidity that cannot be assessed by biopsy alone. This paper demonstrated three different methods to predict microscopic cellular and molecular properties of brain tumors from macroscopic, minimally-invasive clinical images. Elementary statistical evaluations demonstrated that significant relationships between the macroscopic and microscopic variables of interest did exist. Machine learning combined with a conservative correction for family-wise error rates was able to predict cellular and molecular properties with high accuracy but limited classification sensitivity (0.2-2.3%) for three of the four outcome variables. When spatial correlations across voxels were taken into account using Gaussian random field theory, high accuracy was retained with a significant increase in classification sensitivity (3.2-9.9%). The images generated by random field theory demonstrated acceptable noise and spatial resolution properties for clinical interpretation. Taken together, our results show that in vivo microscopic and even genomic mapping of human brain tumors may be clinically possible in the near future. The near-term implication of our findings is that researchers and clinicians utilizing machine learning to predict tumor heterogeneity should consider dimensionality to be one potential vehicle by which in vivo imaging signatures may be used to traverse scale. The rapid expansion of anatomical and functional MR sequences and the growing availability of hybrid imaging systems only serve to enhance this opportunity. The long-term implications of our findings are that it may be possible to map cellular and molecular tumor properties across both space and time during treatment, allowing for highly personalized treatment strategies that are not currently possible.CONCLUSION
We have demonstrated statistical relationships between routine multiparametric imaging signatures and underlying cellular and molecular properties of brain tumors. We have applied advanced statistical methods to correct for the family-wise error rate problem associated with whole-brain statistical parametric mapping, and have shown that the results have strong agreement with surgical biopsy. These results imply that cellular and molecular mapping of tumor heterogeneity from minimally-invasive images may be possible in the near future.Acknowledgements
We thank Ms. Nichole Johnson for her assistance in operating the neuronavigation system, interpreting the coordinate system and uncertainty indices, and exporting the biopsy screen captures. We thank Dr. Anderson Winkler for insight into the application of random field theory to various parametric maps, and specific issues related to implementation of our method using FSL. We thank Dr. Mark Holland for useful conversations related to the use of random field theory to estimate smoothness in the chi-square maps.References
[1] J. J. Olsen, L. Nayak, D. R. Ormond, P. Y. Wen, S. N. Kalkanis and T. C. Ryken, "The role of targeted therapies in the management of progressive glioblastoma," J Neurooncol, vol. 118, pp. 557-99, 2014.
[2] W. Paulus and J. Peiffer, "lntratumoral Histologic Heterogeneity of Gliomas: A Quantitative Study," Cancer, vol. 64, pp. 442-7, 1989.
[3] T. A. Yap, M. Gerlinger, P. A. Futreal, L. Pusztai and C. Swanton, "Intratumor Heterogeneity: Seeing the Wood for the Trees," Sci Transl Med, vol. 4, no. 127, p. 127ps10, 2012.
[4] R. J. Gilles, P. E. Kinahan and H. Hricak, "Radiomics: Images are more than pictures, they are data," Radiology, vol. 278, no. 2, pp. 563-77, 2016.
[5] C. C. Jaffe, "Imaging and genomics: is there a synergy?," Radiology, vol. 264, no. 2, pp. 329-31, 2012.
[6] P. O. Zinn, B. Majadan, P. Sathyan, S. K. Singh, S. Majumder, F. A. Jolesz and R. R. Colen, "Radiogenomic mapping of edema/cellular invasion MRI-phenotypes in glioblastoma multiforme," PLoS one, vol. 6, no. 10, p. e25451, 2011.
[7] M. Diehn, C. Nardini, D. S. Wang, S. McGovern, M. Jayaraman, Y. Liang, K. Aldape, S. Cha and M. D. Kuo, "Identification of noninvasive imaging surrogates for brain tumor gene-expression modules," Proc Natl Acad Sci USA, vol. 105, no. 13, pp. 5213-8, 2008.
Figures