0392
Sodium Relaxometry using Magnetic Resonance Fingerprinting1Medical Physics in Radiology, German Cancer Research Center (DKFZ), Heidelberg, Germany, 2Faculty of Physics and Astronomy, Ruprecht-Karls University Heidelberg, Heidelberg, Germany, 3Physikalisch-Technische Bundesanstalt (PTB), Braunschweig and Berlin, Germany, 4Institute of Radiology, University Hospital Erlangen, Friedrich-Alexander-Universität Erlangen-Nürnberg (FAU), Erlangen, Germany, 5Institute of Medical Physics, Friedrich-Alexander-Universität (FAU), Erlangen, Germany, 6Siemens Healthcare GmbH, Erlangen, Germany, 7Faculty of Medicine, Ruprecht-Karls University Heidelberg, Heidelberg, Germany
Synopsis
Sodium relaxation times have been shown to be altered in several diseases. However, due to short relaxation times and low in-vivo signal, measurement times in sodium relaxometry on the order of 1h were reported for both, longitudinal and transversal relaxation constants. In this work, a novel sodium relaxometry method based on Magnetic Resonance Fingerprinting (MRF) principles is presented, which enables simultaneous quantification of T1, T2s*, T2l*, T2* and ΔB0, with automatic distinction between bi- and monoexponential transverse relaxation.
Introduction
23Na relaxation times in diseased tissue can be altered and relaxation-weighted images can provide additional specific information, e.g. in tumor tissue1,2. Consequently, several studies have aimed for quantification of the relaxation times2,3,4. However, these measurements require long acquisition times due to the low in-vivo concentrations and the low NMR sensitivity of sodium. Magnetic Resonance Fingerprinting (MRF) has been shown to be an efficient technique for multi-parametric quantification in 1H-MRI5. Non-steady-state conditions are generated by temporal variations of excitations, gradients and sequence timings, resulting in a unique signal evolution for each combination of relaxation constants. In this work, we present a technique referred as 23Na-MRF that enables simultaneous quantification of T1, T2s*, T2l*, T2* and ΔB0, with automatic differentiation between bi- and monoexponential transverse relaxation.Methods
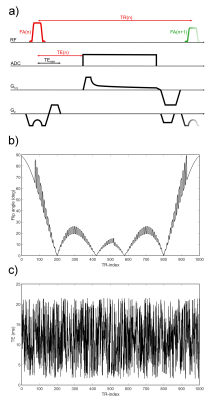
A 2D density-adapted radial spoiled SSFP (FISP) sequence6 using VERSE excitation was used for the acquisition of 1000 timeframes with variable flip angle (FA), echo time (TE) and repetition time (TR) patterns. The sequence scheme and the FA pattern are shown in Fig.1. The TE pattern was chosen pseudo randomly between TEmin=1.55ms and 21.55ms (c.f.Fig.1). In each timeframe the smallest possible TR was chosen; consequently, the TR is equal to the TE plus the durations of the gradients, resulting in a TR pattern between 13.34ms and 33.34ms.For each pulse train, i.e. the measurement of 1 radial spoke in each timeframe, each spoke angle was imcremented by 111.2 degrees relative to the previous one. For a resolution of 2x2x12mm3/4x4x12mm3 14/7 pulse trains were acquired, resulting in 1000 golden angle-rotated timeframes in k-space containing 14/7 spokes each.
In a first step, adapted Bloch equations were assumed to approximate the system, whereby T2s* and T2l* correspond to the short and the long transverse relaxation component:
$$\frac{d}{dt}\begin{pmatrix}M_x\\M_y\\M_z\end{pmatrix}=\begin{pmatrix}-\frac{0.6}{T_{2s}^*}-\frac{0.4}{T_{2l}^*}&{\gamma}B_z&-{\gamma}B_y\\-{\gamma}B_z&-\frac{0.6}{T_{2s}^*}-\frac{0.4}{T_{2l}^*}&{\gamma}B_x\\{\gamma}B_y&-{\gamma}B_x&-\frac{1}{T_{1}}\end{pmatrix}\begin{pmatrix}M_x\\M_y\\M_z\end{pmatrix}+\begin{pmatrix}0\\0\\\frac{M_0}{T_1}\end{pmatrix}$$
Monoexponential transverse relaxation was modeled with T1 and T2*=T2s*=T2l*. Furthermore, deviations from the static field ΔB0 were considered in both models. A dictionary, needed for reconstruction, was simulated containing each combination in the following parameter space: biexponential: T1=[20,21,…,70]ms, T2s*=[1.0,1.3,…,14.8]ms, T2l*=[15,16,…,60]ms, ΔB0=[-60,-58,…,60]Hz; monoexponential: T1=[30,31,…,90]ms, T2*=[5,6,…,80]ms, ΔB0=[-60,-58,…,60]Hz. The resulting dictionary, containing both bi- and monoexponential transverse relaxation, was compressed using an SVD compression (rank 12).
Reconstructions were performed using a low rank alternating direction method of multipliers approach7. An L2 norm in the wavelet domain was used as spatial regularization (λphantom=1*10-2, λinvivo=5*10-2, 40 iterations). The relaxation parameters and ΔB0 were reconstructed pixelwise by finding the highest scalar product between the pixel signal evolution and all dictionary entries. This automatically determined if the bi- or monoexponential model better describes the measured data in each pixel.
Measurements were performed on a 7T research system (Siemens Healthcare, Germany), using a 30-channel 23Na coil with additional 1H channel (Rapid Biomed GmbH, Germany) for in-vivo measurements. Phantom experiments were performed with a single-channel 23Na birdcage coil with 1H channel (Rapid Biomed GmbH, Germany).
As a reference, the longitudinal relaxation constants were determined by fitting a monoexponential model to the data of a 2D density-adapted radial sequence (TR=300ms,FA=63°,2x2x12mm3,TE=1.34ms,10 averages) with a preceding non-selective inversion pulse and varying inversion times TI=[3.2,10,20,40,70,100,130,160,200,250]ms. The transverse relaxation was determined by fitting both a bi- and monoexponential model to the data acquired with the same sequence without the inversion preparation (TR=200ms,FA=60°,2x2x12mm3,10 averages) and varying TE=[1.24,1.35,2.0,2.5,3,4,5,6,7,9,13,17,23,27,35,40,50,60]ms. Each pixel was assigned to bi- or monoexponential relaxation based on the coefficient of determination (R2) of the fits. Reference maps of ΔB0 were determined via phase difference between two data sets acquired with the same sequence (TR=25ms,FA=45°,2x2x12mm3,20 averages) with two different echo times (TE1=1.55ms,TE2=6.55ms).
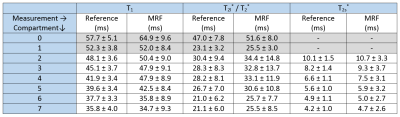
The MRF phantom measurements were performed with a resolution of 2x2x12mm3 (11 averages, 62:16mins measurement time), whereas the in-vivo measurements had a resolution of 4x4x12mm3 (21 averages, 59:26mins measurement time). Mean and standard deviation (SD) were computed in all phantom compartments and in white matter (WM) and cerebrospinal fluid (CSF).
Results
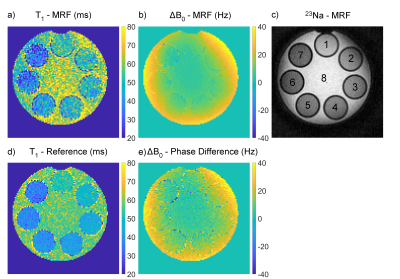
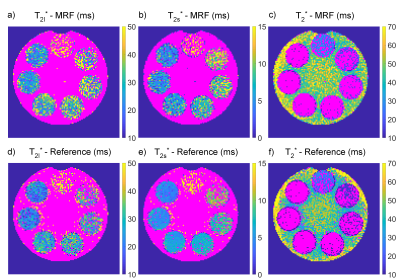
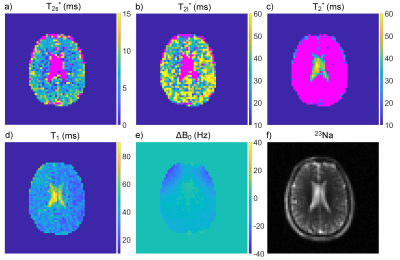
Maps of the relaxation times and ΔB0 in the phantom are shown in Fig.2 and Fig.3. All mean relaxation times acquired with 23Na-MRF are in agreement with the reference results within the SD (c.f.Tab.1).The resulting maps of the in-vivo measurements are displayed in Fig.4. The reconstruction automatically determined biexponential transverse relaxation in brain tissue and monoexponential relaxation in CSF. The following relaxation times, averaged over ROIs, were found for WM: T1=35.1±8.4ms, T2l*=35.2±12.1ms, T2s*=5.0±2.5ms. In CSF the values were: T1=65.5±15.9ms, T2*=42.9±8.0ms.
Discussion and Conclusion
This work shows that 23Na-MRF is highly promising enabling simultaneous measurement of T1, T2s*, T2l*, T2* and ΔB0 in-vivo for the first time.This work is based on adapted Bloch equations as a simplified model that cannot fully describe a spin 3/2 system. This issue could be tackled with irreducible tensor operators8. Furthermore, the relaxation models imply a single tissue compartment per pixel. Another improvement could be the use of more sophisticated FA and TE patterns.
Nevertheless, the mean relaxation times in all phantom compartments acquired with 23Na-MRF are in agreement with the results from the reference method within the error. This demonstrates the potential of 23Na-MRF for 23Na relaxometry. Furthermore, the in-vivo results are in good agreement with literature, where the following values have been reported2,3,4: T1,CSF=64ms, T1,WM=37.1ms, T2,CSF*=56ms, T2l,WM*=25.9-40.9ms, T2s,WM*=1.99-6.5ms.
Acknowledgements
No acknowledgement found.References
1. Kline RP, Wu EX, Petrylak DP, Szabolcs M, Alderson PO, Weisfeldt ML, Cannon P, Katz J. Rapid in vivo monitoring of chemotherapeutic response using weighted sodium magnetic resonance imaging. Clin Cancer Res 2000;6(6):2146-2156.
2. Nagel AM, Bock M, Hartmann C, Gerigk L, Neumann JO, Weber MA, Bendszus M, Radbruch A, Wick W, Schlemmer HP, Semmler W, Biller A. The potential of relaxation-weighted sodium magnetic resonance imaging as demonstrated on brain tumors. Invest Radiol 2011;46(9):539-547.
3. Blunck Y, Josan S, Taqdees SW, Moffat BA, Ordidge RJ, Cleary JO, Johnston LA. 3D-multi-echo radial imaging of (23) Na (3D-MERINA) for time-efficient multi-parameter tissue compartment mapping. Magn Reson Med 2018;79(4):1950-1961
4. Lommen JM, Flassbeck S, Behl NGR, Niesporek S, Bachert P, Ladd ME, Nagel AM. Probing the microscopic environment of (23) Na ions in brain tissue by MRI: On the accuracy of different sampling schemes for the determination of rapid, biexponential T2* decay at low signal-to-noise ratio. Magn Reson Med 2018;80(2):571-584
5. Ma D, Gulani V, Seiberlich N, Liu K, Sunshine JL, Duerk JL, Griswold MA. Magnetic resonance fingerprinting. Nature 2013;495(7440):187-192
6. Kratzer FJ, Flassbeck S, Schmidt S, Nagel AM, Bachert P, Ladd ME, Behl NGR. Density Adapted Stack of Stars Sequence for 23Na Imaging at 7T. In Proceedings of ISMRM Workshop on Imaging of nX-Nuclei. Dubrovnik, Croatia. 2018
7. Asslander J, Cloos MA, Knoll F, Sodickson DK, Hennig J, Lattanzi R. Low rank alternating direction method of multipliers reconstruction for MR fingerprinting. Magn Reson Med 2018;79(1):83-96
8. Van Der Maarel JRC. Thermal Relaxation and Coherence Dynamics of Spin 3/2. I. Static and Fluctuating Quadrupolar Interactions in the Multipole Basis. Concepts in Magnetic Resonance Part A 2003(19A(2)):97-116
Figures