0328
Super-resolution Generative Adversarial Network for improving malignancy characterization of hepatocellular carcinoma1School of Medical Information Engineering, Guangzhou University of Chinese Medicine, Guangzhou, China, 2Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 3Department of Radiology, Guangdong General Hospital, Guangzhou, China
Synopsis
Deep feature derived from data-driven learning has consistently shown to outperform conventional texture features for lesion characterization. However, due to the slice thickness of medical imaging, through-plane has worse resolution than in-plane resolution. Therefore, the performance of deep feature extracted from the through plane slices may be worse, and their contributions to the final characterization may also be very limited. We propose an end-to-end super-resolution and self-attention framework based on generative adversarial network (GAN), in which the through-plane slices with low resolution are enhanced by learning the in-plane slices with high resolution to improve the performance of lesion characterization.
Introduction
The preoperative knowledge of the biological aggressiveness or malignancy degree of hepatocellular carcinoma (HCC) is significant for determining treatment decisions and predicting patient prognosis1. Deep feature based on convolutional neural networks (CNN) has been shown to be promising to represent the biological aggressiveness of neoplasm2-3. However, the slice thickness of MR imaging may remarkably degrade the clarity of 3D lesion images within coronal or sagittal views so as to influence the performance of lesion characterization. To alleviate the problem, we propose an end-to-end super-resolution and classification framework based on Generative adversarial networks (GAN) for improving the malignancy characterization of HCC.Methods
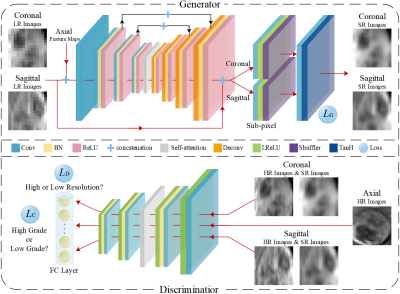
The study was approved by the local institutional review board and the patient's informed consentswere waived. From October 2012 to October 2018, there were 115 cases in 112 subjects (101 males and 11 females, aged 51.09±11.53), ranging from 27 to 79 years) underwent HCC resection. Contrast-enhanced MR images were acquired for all subjects using 3.0 Tesla magnetic resonance scanner (signa excite HD 3.0T, GE health care, milwaukee, wi, usa). The image matrix dimension was 512×512× 92 and the voxel spacing was 0.46mm×0.46mm×2.2mm. Histological grading data for these 115 HCCs were obtained from clinical histological reports confirmed by histopathology, including 3 cases of Edmondson I, 51 cases of Edmondson II, 57 cases of Edmondson III and 4 cases of Edmondson IV. In clinical routine, Edmondson I and II correspond to low-grade and Edmondson III and IV correspond to high-grade. The framework of the proposed method is shown in Figure 1. The whole network consists of two components: a generative module G and a discriminative module D. The super-resolution subnetwork and the classification subnetwork are embedded in the generative module and discriminative module, respectively. The generator is trained with the super-resolution subnetwork to synthesize high-resolution image patches based on the low-resolution input patches corresponding to the coronal and sagittal views. Conversely, the discriminator is trained adversarially to identify whether the synthesized patches are high-resolution or low-resolution. Meanwhile, the classification subnetwork is jointly trained with the discriminator and to decide the high-grade or low-grade of the lesion. Furthermore, a self-attention mechanism4 is adopted to enable both the generator and the discriminator to efficiently capture relationships between widely separated spatial regions, yielding more discriminative features to further improve the performance of super-resolution and classification. The data set is randomly divided into two parts: training and verification set (75 HCCs) and fixed test set (40 HCCs). Receiver operating characteristic curve (ROC) and area under the curve (AUC) were used to assess the characterization performance.Results
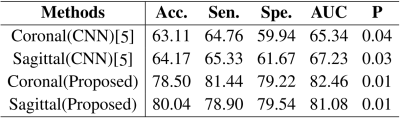
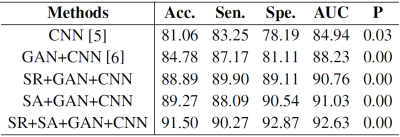
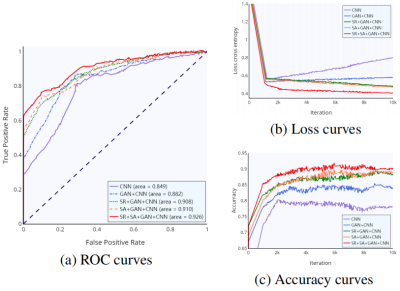
Table 1 showed the performance comparison of through plane patches (coronal or sagittal) for malignancy characterization using the conventional CNN method and the proposed method. It can be clearly observed that the characterization performance of the through-plane slices is rather low with the conventional CNN method5. Comparatively, the proposed method can significantly improve the characterization performance for each view. Table 2 showed the performance comparison of the proposed method and other methods in the conventional framework of multi-view convolutional networks for malignancy characterization of HCC, in which deep features separately extracted from three 2D orthogonal views by CNN were concatenated for classification. It can be found that the GAN+CNN method6 is superior to the traditional CNN method due to the adoption of GAN for more sample generation. Subsequently, the proposed method with the super-resolution (SR) module can significantly improve the characterization performance. Furthermore, the proposed method with the adoption of self-attention (SA) also obtain further improved performance. The ROC curves, loss curves, and accuracy curves of the performance comparison corresponding to Table 2 were plotted in Fig. 2.Discussion
The proposed method can signifificantly improve the characterization performance for each view. There might be several reasons accounting for these improvements. On the one hand, the proposed method can yield more high-resolution samples so as to alleviate the problem of over-fitting due to the limited training data. On the other hand, the proposed method can extract multiple features from widely separated regions in high-resolution images for performance improvement of characterization. Subsequently, the proposed method with the super-resolution module can significantly improve the characterization performance. This is attributed to the adoption of super-resolution, which can make the GAN generate more high-resolution images. Furthermore, the proposed method with the adoption of self-attention also shows that further improved performance can be obtained because of the aggregation of multiple features from widely separated regions. Note that the loss curve of the conventional CNN method has a unwarping shape, and such slight over-fitting might be caused by the limitation of training data. Comparatively, the proposed method can remarkably alleviate the risk of over-fitting due to the adoption of GAN with super-resolution and self-attention to generate more high-quality samples.Conclusion
An end-to-end super-resolution and classification framework based on conditional adversarial networks (CAN) is proposed to alliviate the problem of slice thickness of MR for improving the malignancy characterization of HCC. Experimental results of clinical HCCs demonstrate the superior performance of the proposed method compared with other methods. Our experiments for MRI super-resolution and classification also give insight to design proper network architectures for lesion characterization in clinical practice.Acknowledgements
This research is supported by the grant from National Natural Science Foundation of China (NSFC: 81771920).References
[1] Sasaki A, Kai S, Iwashita Y, et al. Microsatellite distribution and indication for locoregional therapy in small hepatocellular carcinoma. Cancer 2005;103( 2): 299-306.
[2] Ciompi F, de Hoop B, van Riel SJ, et al. Automatic classification of pulmonary peri-fissural nodules in computed tomography using an ensemble of 2D views and a convolutional neural network out-of-the box. Med. Imag. Anal.2015;26(1):195-202.
[3] Roth H, Lu L, Liu J, et al. Improving computer-aided detection using Convolutional Neural Networks and Random View Aggregation. IEEE Trans. Med. Imag. 2016;35(5):1170-1181.
[4] Zhang H, Goodfellow I, Metaxas D, Odena A. Self-Attention Generative Adversarial Network. arXiv:1805.08318, Jun. 2019.
[5] Wang Q, Zhang L, Xie Y, Zheng H, Zhou W. Malignancy characterization of hepatocellular carcinoma using hybrid texture and deep feature. Proc. IEEE 24th Int. Conf. Image Process (ICIP), Sep. 2017: 4162-4166.
[6] Frid-Adar M, Klang E, Amitai M, Goldberger J, Greenspan H. Synthetic data augmentation using GAN for improved liver lesion classification. Proc. IEEE 15th Int. Symp. Biomed. Imag. (ISBI), Apr. 2018: 289-293.
Figures