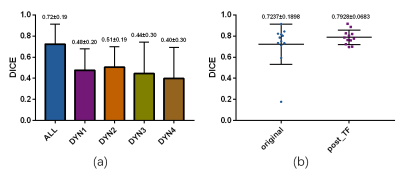0305
Automatic Liver Tumor Detection using Deep learning based segmentation and Radiomics guided Candidate Filtering1Institute of Science and Technology for Brain-Inspired Intelligence, Fudan University, Shanghai, China, 2Radiology department, The first affiliated hospital, College of medicine, Zhejiang University, Hangzhou, China, 3Human Phenome Institute, Fudan University, Shanghai, China
Synopsis
The objective of this study is to perform automatic multi DCE phases liver tumor detection using deep learning based segmentation and radiomics guided candidate filtering. The proposed model consists mainly of two stages, primary segmentation based on a U-net architecture neural network in stage1, and suspected tumor discrimination mechanism using multi DCE phases radiomics features including shape features, texture features, time dimension features and location information in stage 2. The proposed two-stage model exhibits superior performance in HCC tumor segmentation with a mean Dice score of 0.7928 in test set.
INTRODUCTION
Liver cancer is one of the most common cause of cancer-induced deaths according to the statistic from World Health Organization. Hepatocellular carcinoma (HCC) as the most common type of primary liver cancer, which is the sixth most prevalent cancer1. Early diagnosis and treatment for HCC are crucial for improving the success rate of tumor resection and survival rate2. Accurate tumor segmentation is an important prerequisite for quantitative analysis and evaluation of tumors, which can help doctors better understand tumors and develop appropriate treatment plans. Magnetic Resonance (MR) Dynamic Contrast Enhanced (DCE) imaging is a common imaging modality in liver cancer, this study proposed a model combining multi DCE phases to address the liver tumor detection task through deep learning based segmentation and radiomics features guided candidate filtering.METHODS
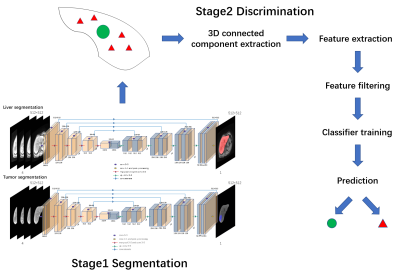
95 HCC patients were recruited into the subject group for gadolinium-based contrast agent enhanced MR scans. Four DCE phases (mask phase, arterial phase, portal venous phase, delayed phase) were acquired for each participate. The whole patient set was randomly divided into train set (71 cases, for neural network and discriminator training), validation set (12 cases, for evaluation and hyperparameter adjustment) and test set (12 cases, for generalization ability evaluation). The liver tumor detection was mainly divided into two stages, the first stage is primary segmentation, in which (a) a classical 2D U-net architecture neural network with ‘batch normalization’ was adopted for liver segmentation. The input of the network was 4-channel co-registered DCE phases, where the registration was based on Symmetric Normalization (SyN) algorithm3, performed in Advanced Normalization Tools (ANTs). (b) Areas outside the liver were regarded as background and would not be considered in subsequent tumor segmentation. (c) The segmented liver ROI was used as the input of the second neural network for tumor segmentation. Tumor candidates generated after a relatively small threshold (0.3 in practice) in tumor segmentation probability map to ensure that most suspected tumor areas were included in. The second stage was suspected tumor candidates filtering based on radiomics features. (d) Each suspected tumor area was identified as a 3D connecting component for radiomics features extraction, texture features and common shape features were extracted in four DCE phases. Beyond that some time dimension features were extracted as well, including the difference of texture features on the timeline and time curve features (kurtosis, skewness, mean and variance). Additionally, there were two other features describing the location of the tumor candidate. In general, 1108 radiomics features were included in the feature set. (e) The least absolute shrinkage and selection operator (LASSO) logistic regression4 was applied to do the feature selection, and the top ten essential elements were used in the classification model. (f) Various classifiers were trained and evaluated in the 5-fold cross-validation, the optimal classifier was chosen to predict in the test set. After the two-stage segmentation and discrimination, the identified HCC tumor components were regarded as the final results. 3D volume Dice similarity coefficient (Dice) score was adopted to evaluate the segmentation performance. The whole framework is shown in Figure 1.RESULTS
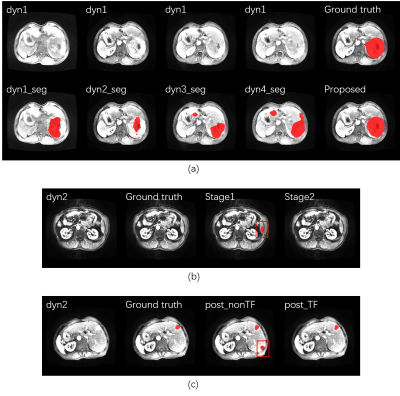
The liver segmentation achieved mean Dice score of 0.9610 in test set. The primary tumor segmentation based on multi DCE phases in the first stage achieved mean Dice score of 0.7237, which was more dominant than tumor segmentation based on one DCE phase (Figure 2 (a)). After the second stage discrimination using radiomics features, the final mean Dice score improved to 0.7928 (Figure 2 (b)). Compared to discrimination mechanism without time dimension features, the proposed model had a better discrimination performance, manifested in accuracy, area under curve (AUC) value, F1 score, false positive rate (FPR) and final segmentation Dice scores (Figure 3). A segmentation slice demonstration is illustrated in Figure 4.DISCUSSION
The results indicated that the proposed automatic two-stage segmentation model has a superior performance in liver tumor segmentation compared to some existed methods5, 6. High false positive rate is common for small region detection after primary segmentation through neural network, hence a discrimination mechanism was adopted to filter the ‘fake tumor’ for a better segmentation performance. Typically, shape features were considered important in discrimination. However, due to the relatively poor segmentation results in test set compared to training set, the shape characteristics might be destroyed and show a weakened influence in discrimination. In this study, time dimension features in multi DCE phases were extracted and considered to be helpful for true tumors identification. The reason was that due to the influence of contrast agent, the response of tumor region in multi DCE phases was different from external areas. This difference could be reflected in the time dimension features and generally not affected by the segmentation effect, resulted in a better discrimination performance.CONCLUSION
This study demonstrated an automatic two-stage liver tumor segmentation model, which consists of a primary segmentation model based on deep learning and a discrimination mechanism guided by radiomics features. The proposed model had a considerable performance in HCC tumor segmentation.Acknowledgements
Acknowledge: This work was supported by Shanghai Municipal Science and Technology Major Project (No.2017SHZDZX01), Shanghai Municipal Science and Technology Major Project (No.2018SHZDZX01) and ZJLab, Shanghai Natural Science Foundation (No. 17ZR1401600) and the National Natural Science Foundation of China (No. 81971583).References
1. Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet (London, England) 2012;379:1245-1255.
2. Radtke A, Nadalin S, Sotiropoulos GC, et al. Computer-assisted operative planning in adult living donor liver transplantation: a new way to resolve the dilemma of the middle hepatic vein. World journal of surgery 2007;31:175-185.
3. Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal 2008;12:26-41.
4. Tibshirani R. Regression Shrinkage and Selection via the Lasso. Journal of the Royal Statistical Society Series B (Methodological) 1996;58:267-288.
5. Chlebus G, Meine H, Moltz J, Schenk A. Neureal Network-Based Automatic Liver Tumor Segmentation With Random Forest-Based Candidate Filtering. 2017.
6. Grzegorz Chlebus AS, Jan Hendrik Moltz. Deep learning based automatic liver tumor segmentation in CT with shape-based post-processing. 2018: Unpublished.
Figures