0244
Deep-learning Diagnosis of Supraspinatus Tendon Tears: Comparison of Multi-sequence Versus Single Sequence Input1Department of Radiology, NYU School of Medicine, New York, NY, United States, 2Siemens Healthineers, Princeton, NJ, United States, 3Siemens Healthineers, Erlangen, Germany
Synopsis
Rotator cuff tears are a common cause of shoulder pain and typically diagnosed on shoulder MRI. Using 1,218 MR examinations performed at multiple field strengths and from multiple vendors, we developed a deep-learning (DL) model for the diagnosis of supraspinatus tendon tears on MRI using an ensemble of 3D ResNets combined via logistic regression to classify tears into no tear, partial tear, and full-thickness tear. We compared the effect of using multiple sequences as input versus a single sequence. Our results show that deep-learning diagnosis of supraspinatus tendon tears is feasible and that multi-sequence input improves model performance.
Introduction
Rotator cuff tears are a common cause of shoulder disability, affecting nearly 40% of individuals older than 60 years and over half of individuals older than 80 years.1 MRI is widely used for rotator cuff tear detection and characterization to help guide surgical versus non-surgical management. Radiologist interpretation of rotator cuff tears can be time-consuming and consists of tear classification (partial versus full-thickness) for each tendon as well as quantitative measurements and qualitative descriptions of muscle and tendon status.2DL has the potential to aid radiologists by increasing both their accuracy3 and productivity. Although all tendons of the rotator cuff may tear, the supraspinatus tendon is the most commonly torn tendon. The purpose of this study was to develop a DL model for the diagnosis of supraspinatus tendon tears on shoulder MRI. In addition, we compared the effect of using a single MRI sequence versus multiple sequences as input for the DL model.
Methods
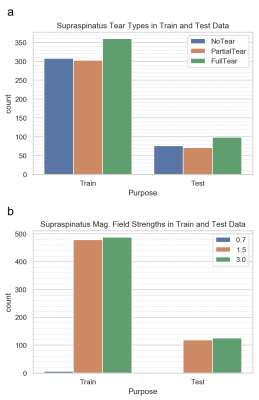
This was a HIPAA-compliant, IRB-approved study. The dataset consisted of 1,218 MRIs obtained on Philips, Siemens, GE, Toshiba, and Hitachi scanners at 0.7, 1.5 or 3T. Each examination consisted of coronal, sagittal, and axial fluid-sensitive (FS) (T2-weighted fat-suppressed, proton density weighted fat-suppressed, or STIR) plus sagittal T1-weighted sequences (Figure 2). Radiology reports were used as ground truth for tear classification, which was defined as no tear, partial tear, or full-thickness tear.The data was split into 972/246 for training/testing, limiting the test cases to 1.5 and 3T only, due to the small number of 0.7T cases. Distribution of tear types and magnetic field strengths on the training and test data are shown in Figure 1. All images were preprocessed by linear scaling of the intensity values to the range [0,1]. All left shoulder images were flipped to simulate right shoulder views. Images were then resampled to a fixed image size of 128, 128, 32 (x,y,z) using trilinear interpolation.
The classification stage comprised 4 parallel 3D ResNet50 CNN architectures4 followed by a logistic regression (Figure 3). The ResNet50 models were trained via transfer learning.5 They were initialized with weights pretrained on the Kinetics dataset6,7 and then further trained on the 972 training cases to adapt the models to the targeted domain. Each of the ResNet50 models was independently trained on a particular view out of the four views. Training was set up with Adam optimization, cross-entropy loss, and a learning rate of 10-5. A logistic regression was trained to weight the independent per-view predictions of the 4 models to combine them into the final prediction.
A receiver operating characteristic (ROC) analysis was performed to assess the performance of the full classification pipeline versus using each MRI sequence alone, and the influence of magnetic field strength on the classification results.
Results
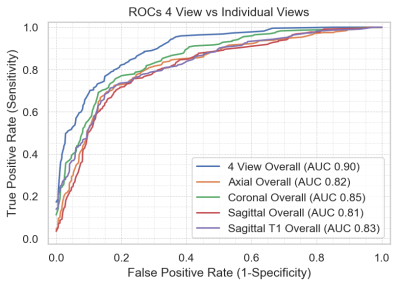
The area under the curve (AUC) for the multi-sequence input was 0.90, which was higher than each of the individual single sequences tested (0.81-0.85). Figure 4 shows the comparison between the multi-sequence and single sequence input. Of the single sequence input, the coronal FS sequence demonstrated the highest AUC. Among the different tear types, the model performed better for full-thickness tears, followed by no tears and partial-thickness tears (Figure 5). The model performed better at 3T versus 1.5T, with AUC 0.91 versus 0.90, respectively.Discussion
We have demonstrated that deep-learning diagnosis of supraspinatus tendon tears is feasible with reasonable diagnostic performance. As with human readers, model performance improved with the use of multiple sequences compared to a single sequence. Although this confirms our hypothesis, the initial rationale to investigate whether the model could diagnose tendon tears on a single sequence was based on the idea that if the model could in fact perform well with a single sequence alone, this would contribute further to simplification of the classification task and potentially decrease image acquisition time as well. Our results demonstrate that multi-sequence image acquisition is needed for adequate model performance.The comparison among the single sequence inputs shows that the model performs best with the coronal FS sequence. This is in concordance with clinical practice, where radiologists rely heavily on this sequence to assess the integrity of the supraspinatus tendon. Interestingly, the model performs slightly better with the axial FS sequence when compared to the sagittal FS sequence. Typically, the sagittal FS sequence would be considered by most radiologists to be the other sequence they rely upon for accurate diagnosis of supraspinatus tendon tears, apart from the coronal FS sequence.
The comparison among tear types shows that the model performs best with full-thickness tears, followed by no tears and partial-thickness tears. The lower performance for partial-thickness tears for both the model as well as human readers corroborates the known challenges and reader variability for partial tears. The superior performance of the model for full-thickness tears compared to no tears may be due to the discrete fluid signal that can be more easily identified with full-thickness tears, whereas patients without tears may have varying degrees of signal abnormality and heterogeneity due to varying degrees of tendinosis.
Conclusion
Deep-learning diagnosis of supraspinatus tendon tears is feasible. Multi-sequence input improves tear classification compared to single-sequence.Acknowledgements
We acknowledge Heiko Meyer for his valuable input and support for this project. We acknowledge Shivam Kausik for annotation/labeling assistance.References
1. Tashjian RZ. Epidemiology, natural history, and indications for treatment of rotator cuff tears. Clin Sports Med. 2012;31(4): 589-604.
2. Morag Y, Jacobson JA, Miller B, et al. MR imaging of rotator cuff injury: what the clinician needs to know. Radiographics. 2006;26(4):1045-65.
3. Bien N, Rajpurkar P, Ball RL, et al. Deep-learning-assisted diagnosis for knee magnetic resonance imaging: Development and retrospective validation of MRNet. PLOS Medicine. 2018;15(11): e1002699. https://doi.org/10.1371/journal.pmed.1002699
4. He K, Zhang X, Ren S, et al. (2016). Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 770-778).
5. Yosinski J, Clune J, Bengio Y, et al. (2014). How transferable are features in deep neural networks?. In Advances in neural information processing systems (pp. 3320-3328).
6. Hara K, Kataoka H, Satoh Y. (2018). Can Spatiotemporal 3D CNNs Retrace the History of 2D CNNs and ImageNet?. In Proceedings of the IEEE conference on Computer Vision and Pattern Recognition (pp. 6546-6555).
7. Kay W, Carreira J, Simonyan K, et al. (2017). The kinetics human action video dataset. arXiv preprint arXiv:1705.06950.
Figures