0243
Semi-Quantitative Grading of the Anterior Cruciate Ligament using Deep Learning1Department of Radiology and Biomedical Imaging, University of California, San Francisco, San Francisco, CA, United States
Synopsis
In this study we present a fully-automated anterior cruciate ligament (ACL) detection and classification framework which provides multi-class severity staging of ACL tears using state-of-the-art deep learning architectures. We compared the performances of a 3D and a 2D convolutional neural network (CNN) in ACL lesion classification. A higher overall accuracy (84%) and linear-weighted kappa (.92) were observed with the 2D model; however, it underperformed compared to the 3D CNN in classifying partial tears. This is the first reported deep learning detection and classification pipeline for ACL severity staging, including reconstructed, fully torn, partially torn, and intact ligaments.
Introduction
The anterior cruciate ligament (ACL) is the most commonly injured ligament in the knee.1,2 ACL injuries increase the risk of developing posttraumatic knee osteoarthritis (OA) and total knee replacement.3–6 MRI is the most effective imaging modality for distinguishing structural properties and abnormalities of the ACL in relation to adjacent musculoskeletal structures.7–10 Several multi-grading scoring systems have been developed to standardize reporting of knee joint abnormalities using MRI;11,12 however, they are susceptible to inter-rater variability, especially if tears are incomplete or there is associated mucoid degeneration.10,13In this study, we propose a deep learning-based pipeline to isolate the ACL region of interest in the knee, detect ACL abnormalities, and stage lesion severity using novel three-dimensional (3D) and previously reported two-dimensional (2D) convolutional neural networks (CNNs). We compare the 3D model with the state-of-the-art 2D network. Additionally, to the best of our knowledge this is the first instance of multi-class ACL severity staging using deep learning, in which reconstructed, fully torn, partially torn, and intact ACLs are graded in accordance with semi-quantitative scoring. This deep learning pipeline would lend towards standardization and generalizability in assessing ACL lesions if deployed clinically.
Methods
A total of 1252 knee MRI studies (202 unique subjects) were obtained from three prior research studies aimed to study joint degeneration in OA and after ACL injury. A 3D fast spin-echo CUBE-sequence was used in all imaging studies with parameters shown in Table 1A.Between 2011 and 2014, five board-certified radiologists (each with over 5 years of training) graded a non-overlapping section of the dataset. The ACL was graded analogous to the WORMS and ACLOAS scoring metrics and included intact, partially torn, fully torn, and reconstructed ACLs. Distribution of gradings and training/validation/test sets splits (70%/10%/20%) are demonstrated in Table 1B.
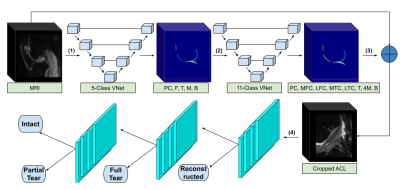
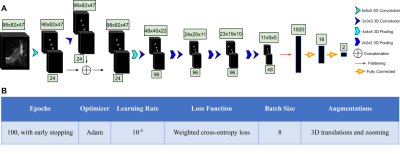
Our framework consisted of two V-Net encoder-decoder architectures that segmented and labeled 11 distinct anatomical components (6 cartilage compartments and 4 meniscus), followed by a rule-based image cropping to isolate the ACL, and a hierarchical tree of CNNs to classify lesion severity (Figure 1). This approach was chosen over a classical multi-class approach because of the non-independence of the classes. The ACL volumes were input into a CNN consisting of 3D convolutional kernels.9 The network was built with six layers, including one skip connection after the first convolution, to preserve initial features and mitigate overfitting (Figure 2A). Performance of the 3D CNN was compared with state-of-the-art 2D CNN MRNet.14 In the MRNet, each slice of the input 3D volume is passed through an AlexNet to extract features.15 The MRNet was pre-trained on the ImageNet dataset and additionally trained with the same training sets and hyperparameters as the 3D CNN (Figure 2B) to ensure correct comparison.
Results
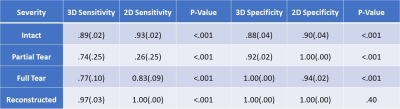
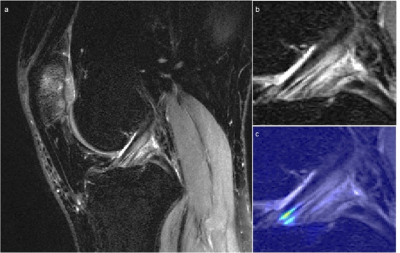
Using the radiologists’ assessment as a standard of reference, the 3D CNN and 2D CNN had overall accuracies (±SD) of 82%±4% and 84%±4% (p=.005), and linear-weighted kappas of .88±.02 and .92±.02 (p<.001), respectively when evaluated on the hold out test set. However, the 2D model correctly classified only one of the four partial tears in the test set, whereas the 3D CNN correctly classified three of the four. Moreover, of the misclassified intact ligaments by the 3D CNN, 94% were misclassified as the adjacent partial tear class, whereas the 2D CNN misclassification of intact ACLs occurred in the full tear class. Both models performed highest on reconstructed ACL classification.The 2D CNN demonstrated higher sensitivity and specificity in detecting intact ACLs (Table 2). The 3D CNN surpassed the 2D CNN in partial tear sensitivity and full tear specificity, whereas the 2D CNN performed better in partial tear specificity and full tear sensitivity. The sensitivity of the 2D CNN in reconstructed ACLs was higher than that of the 3D CNN. The specificities in reconstructed classification were not significantly different. Figure 3 displays a knee with intact ACL that was input into the pipeline, followed by localization of the ACL and the corresponding saliency map generated by the model’s classification weighting.
Discussion
Compared to prior deep learning studies with the ACL,14,16,17 our fully automated pipeline uses 3D convolutional kernels to semi-quantitatively stage the ACL into intact, partially torn, fully torn, and reconstructed. Overall, the performance of the 3D CNN did not surpass that of the 2D model, although the latter was unable to accurately classify partial tears. The global pooling of the MRNet’s slice-by-slice approach may have been unable to capture the subtle differences in a relatively small proportion of slices. The 2D CNN also utilized transfer learning with weight freezing, which may have led to convergence at a local minimum that was closer to the initialized weights but not necessarily highly accurate.Conclusion
Both CNNs evaluated in this study offer distinct benefits; a 2D CNN shows greater overall performance, while the 3D model without any transfer learning can learn more detailed features present in complex gradings. Both architectures displayed a relatively high degree of sensitivity and specificity for intact, fully torn, partially torn, and reconstructed ACLs, which may warrant clinical value of deep learning as a tool for standardizing and generalizing ACL grading.Acknowledgements
No acknowledgement found.References
1. Spindler KP, Wright RW. Anterior cruciate ligament tear. N Engl J Med. 2008;359(20):2135-2142.
2. Johnston JT, Mandelbaum BR, Schub D, et al. Video analysis of anterior cruciate ligament tears in professional American football athletes. Am J Sports Med. 2018;46(4):862-868.
3. Hunter DJ, Lohmander LS, Makovey J, et al. The effect of anterior cruciate ligament injury on bone curvature: exploratory analysis in the KANON trial. Osteoarthr Cartil. 2014;22(7):959-968.
4. Prodromos CC, Han Y, Rogowski J, Joyce B, Shi K. A meta-analysis of the incidence of anterior cruciate ligament tears as a function of gender, sport, and a knee injury–reduction regimen. Arthrosc J Arthrosc Relat Surg. 2007;23(12):1320-1325.
5. Brophy RH, Gill CS, Lyman S, Barnes RP, Rodeo SA, Warren RF. Effect of anterior cruciate ligament reconstruction and meniscectomy on length of career in National Football League athletes: a case control study. Am J Sports Med. 2009;37(11):2102-2107.
6. Suter LG, Smith SR, Katz JN, et al. Projecting lifetime risk of symptomatic knee osteoarthritis and total knee replacement in individuals sustaining a complete anterior cruciate ligament tear in early adulthood. Arthritis Care Res (Hoboken). 2017;69(2):201-208.
7. Shakoor D, Guermazi A, Kijowski R, et al. Cruciate ligament injuries of the knee: A meta‐analysis of the diagnostic performance of 3D MRI. J Magn Reson Imaging. 2019.
8. Ai T, Zhang W, Priddy NK, Li X. Diagnostic performance of CUBE MRI sequences of the knee compared with conventional MRI. Clin Radiol. 2012;67(12):e58-e63.
9. Pedoia V, Norman B, Mehany SN, Bucknor MD, Link TM, Majumdar S. 3D convolutional neural networks for detection and severity staging of meniscus and PFJ cartilage morphological degenerative changes in osteoarthritis and anterior cruciate ligament subjects. J Magn Reson Imaging. 2019;49(2):400-410.
10. Li K, Du J, Huang L-X, Ni L, Liu T, Yang H-L. The diagnostic accuracy of magnetic resonance imaging for anterior cruciate ligament injury in comparison to arthroscopy: a meta-analysis. Sci Rep. 2017;7(1):7583.
11. Hunter DJ, Guermazi A, Lo GH, et al. Evolution of semi-quantitative whole joint assessment of knee OA: MOAKS (MRI Osteoarthritis Knee Score). Osteoarthr Cartil. 2011;19(8):990-1002.
12. Brandt KD, Fife RS, Braunstein EM, Katz B. Radiographic grading of the severity of knee osteoarthritis: relation of the Kellgren and Lawrence grade to a grade based on joint space narrowing, and correlation with arthroscopic evidence of articular cartilage degeneration. Arthritis Rheum. 1991;34(11):1381-1386.
13. Crawford R, Walley G, Bridgman S, Maffulli N. Magnetic resonance imaging versus arthroscopy in the diagnosis of knee pathology, concentrating on meniscal lesions and ACL tears: a systematic review. Br Med Bull. 2007;84(1):5-23.
14. Bien N, Rajpurkar P, Ball RL, et al. Deep-learning-assisted diagnosis for knee magnetic resonance imaging: development and retrospective validation of MRNet. PLoS Med. 2018;15(11):e1002699.
15. Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. In: Advances in Neural Information Processing Systems. ; 2012:1097-1105.
16. Chang PD, Wong TT, Rasiej MJ. Deep Learning for Detection of Complete Anterior Cruciate Ligament Tear. J Digit Imaging. 2019:1-7.
17. Liu F, Guan B, Zhou Z, et al. Fully Automated Diagnosis of Anterior Cruciate Ligament Tears on Knee MR Images by Using Deep Learning. Radiol Artif Intell. 2019;1(3):180091.
Figures