0169
OSCILLATE: A Low-Rank Approach for Accelerated Magnetic Resonance Elastography1Biomedical Engineering, University of Delaware, Newark, DE, United States, 2Bioengineering, University of Illinois, Urbana, IL, United States, 3Biomedical Imaging Research Institute, Cedars-Sinai Medical Center, Los Angeles, CA, United States, 4Thayer School of Engineering, Dartmouth College, Hanover, NH, United States
Synopsis
MR elastography (MRE) has emerged as a sensitive measure of brain health, but due to the need for repeated measures of wave motion, it is a fundamentally long scan. We have developed a method for accurate MRE using spatiotemporal undersampling and low rank joint reconstruction across all samples. We demonstrate the ability to collect accurate MRE data in half the time with under 2% stiffness error. This accelerated method will be used to scan challenging populations, such as those with developmental disabilities, as well as improve achievable resolution and feasibility of multi-frequency or multi-excitation methods.
Introduction
Magnetic resonance elastography (MRE) is a technique for measuring brain tissue mechanical properties to characterize microstructural integrity1. Due to the need to capture tissue deformation in multiple directions over time, MRE is an inherently long acquisition, which limits achievable resolution, multi-frequency or multi-excitation methods, and use in potentially challenging populations, such as children. We have previously proposed a theory for accelerating MRE acquisition by using low-rank image reconstruction to exploit spatiotemporal correlations2, however the previous study only retrospectively undersampled data and exhibited unsatisfactory error metrics owing to an inadequate base sequence. Here we prospectively implement this low-rank technique, called OSCILLATE, along with parallel imaging in an advanced MRE sequence and demonstrate that whole-brain MRE at 2 mm resolution is achievable in under two minutes.Theory
MRE data can be considered low-rank in the sense that a reduced number of spatial and temporal basis functions can describe the entire dataset3. MRE data uses motion-encoding gradients applied separately in three orthogonal directions, with positive and negative gradient polarities, and four equally-spaced samples across one period of harmonic vibration. Therefore, the entire MRE dataset is an effective timeseries of 24 repetitions which are a single magnitude with different phase. As the images are from the same harmonic motion, this leads to a highly spatiotemporally redundant data set, which can be modeled as a partially-separable function in image space described by its Casorati matrix, $$$C=\PsiΦ$$$ , having rank L (< 24): $$\rho(r,\tau)=\sum _{L}^{l=1}\psi_{l}(r)\varphi _{l}\tau $$ with $$$\Psi_{ij} = \psi_{j}(r_{i})$$$ as spatial coefficient maps and $$$\Phi_{ij} = \varphi _{j}(r_{i})$$$ as temporal basis components.Using this model for reconstruction, $$$\Psi$$$ can be estimated if $$$\Phi$$$ is known, and $$$\Phi$$$ can be found from singular value decomposition (SVD) of navigator images acquired with each k-space shot, which have the same contrast and phase as associated imaging data, but lower spatial resolution. The matrix of spatial coefficients $$$\Psi$$$ is iteratively solved for using the objective function: $$\hat{\Psi}=argmin_{\Psi }\left \| d(k,\tau)-E{(\Psi \Phi)} \right \|_{2}^{2} + R{(\Psi) }$$ where d(k,τ) is the acquired k-space data, E is the sampling operator, and R is a spatial roughness penalty.
OSCILLATE allows for reconstruction from undersampled data in a multishot MRE sequence. The proposed OSCILLATE sampling scheme involves alternating which k-space shots are sampled between each repetition, thus achieving an effective reduction factor Rosc. Navigator images are still acquired at each repetition to estimate $$$\Phi$$$ for reconstruction. As all k-space locations are still sampled (though distributed across repetitions), OSCILLATE is separate from, and complementary to, parallel imaging. Figure 1 illustrates the OSCILLATE acquisition and reconstruction scheme.
Method
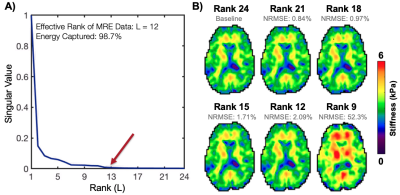
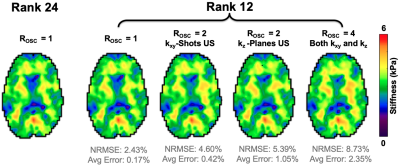
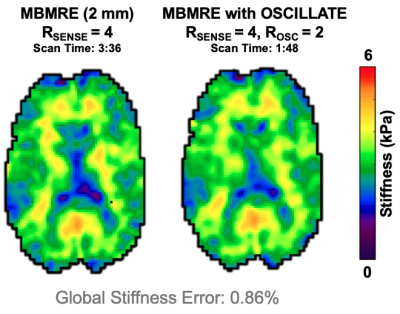
Three MRE data sets from healthy young adults (1M/2F) were acquired on a Siemens 3T Prisma scanner with 64-channel head coil using a 3D multiband, multishot spiral MRE sequence (MBMRE) with parallel imaging RSENSE=4 (Rxy=2 and Rz=2)4. All acquisitions had 2.0x2.0x2.0 mm3 resolution and MRE was performed with 50 Hz vibrations.Displacement data was processed through a nonlinear inversion algorithm5 (NLI) to solve for the complex shear modulus, G, which was converted to viscoelastic shear stiffness, μ=2|G|2/(G’+|G|). Three analyses were conducted to validate the OSCILLATE approach. For each analysis, NRMSE of the shear stiffness map was found, with the fully-sampled, full-rank (L=24) scan as the comparison. Analysis 1: To demonstrate inherent rank of MRE data, SVD of full datasets was conducted after image reconstruction, and data was truncated to various ranks prior to inversion. Analysis 2: Fully-sampled data was retrospectively undersampled and reconstructed with OSCILLATE at low-rank L=12. Sampling patterns included (1) Rosc=2 undersampling of kxy-shots; (2) Rosc=2 undersampling of kz-planes; and (3) Rosc=4 undersampling in both kxy and kz. Analysis 3: Prospectively undersampled data was acquired with Rosc=2 (kxy-shots) and reconstructed with OSCILLATE for comparison with the original MBMRE sequence.Results and Discussion
SVD of MRE displacement data demonstrated that MRE data is accurately described at reduced rank of L=12, compared to the full-rank of 24 (comprising directions, polarities, and timepoints). Nearly all energy is captured at or below L=12 (98.3±0.45%), and the resulting stiffness maps from truncation to L=12 are highly accurate with NRMSE of 2.23±0.11% (Figure 2). It can be assumed that data in higher orders is comprised predominantly of noise, and not reconstructing this data will lead to faster scan times without loss of SNR.Retrospectively undersampling data and reconstructing at low-rank (L=12) similarly yields highly accurate results. Both Rosc=2 undersampling patterns performed well, but undersampling only the kxy-shots achieved the lowest NRMSE (4.60% vs. 5.39%) and global stiffness error (0.42% vs. 1.05%). Interestingly, Rosc=4 still produced very good stiffness maps (global error 2.35%) suggesting the potential to further accelerate with additional regularization (Figure 3). Prospectively undersampled OSCILLATE data sets with Rosc=2 were successfully reconstructed without loss in material property map integrity (average whole-brain error 1.35%; average NRMSE 10.5%; Figure 4).
Conclusion
Due to spatiotemporally redundant information, MRE data can be modeled as low-rank and undersampled data can be reconstructed using OSCILLATE. We show that OSCILLATE is complementary to parallel imaging and can be added for acceleration without loss in image integrity. MBMRE with OSCILLATE produces whole-brain data at 2 mm resolution in 1 minute 48 sec, which is currently the fastest achievable MRE scan time at that resolution.Acknowledgements
NIH/NIBIB R01-EB027577, NIH/NIA R01-AG058853,Delaware INBRE P20-GM103446, and University of Delaware Research Foundation.References
1. Hiscox L V, Johnson CL, Barnhill E, et al. Magnetic resonance elastography (MRE) of the human brain : technique , findings and clinical applications. Phys Med Biol. 2016;61:R401-R437. doi:10.1088/0031-9155/61/24/R401
2. Johnson CL, Holtrop JL, Christodoulou AG, et al. Accelerating MR Elastography with Sparse Sampling and Low-Rank Reconstruction. In: 22nd Annual Meeting of the International Society for Magnetic Resonance in Medicine, Milan, Italy, 2014: p. 325
3. Liang Z. Spatiotemporal Imagingwith Partially Separable Functions. 2007 4th IEEE Int Symp Biomed Imaging From Nano to Macro. 2007;2:988-991. doi:10.1109/ISBI.2007.357020
4. Johnson CL, Holtrop JL, Anderson AT, Sutton BP. Brain MR Elastography with Multiband Excitation and Nonlinear Motion-Induced Phase Error Correction. In: 24th Annual Meeting of the International Society for Magnetic Resonance in Medicine, Singapore, 2016: p. 1951
5. McGarry MDJ, Houten EEW Van, Johnson CL, et al. Multiresolution MR elastography using nonlinear inversion. Med Phys. 2012;39(6388):6388-6396. doi:10.1118/1.4754649
Figures