0162
Fast Whole-Brain MR Elastography Using 3D GRE Multishot Variable Density Spiral Staircase Acquisition1Department of Radiology, Mayo Clinic, Rochester, MN, United States, 2Philips Healthcare, Gainesville, FL, United States
Synopsis
This work reports a new method for fast whole-brain MRE using a 3D gradient-echo multishot variable-density spiral staircase acquisition. The proposed method enables high-resolution MRE by exploiting the inherently improved through-plane parallel imaging. The displacement-SNR is further enhanced by exploiting a motion encoding gradient of a 2 wave-period duration. Constrained reconstruction and deblurring method are used to generate high-quality spiral images. By integrating all these features, the proposed technique provides flexible trade-off among SNR, resolution, spatial blurring and scan time. In vivo experiments have demonstrated the capability of the proposed method for high-SNR high-resolution MRE in less than 5 minutes.
INTRODUCTION
MR elastography has been recognized as a useful tool in studies of neural degenerative disease in the brain1-3. However, resolving accurate tissue mechanical property from in vivo MRE scans requires high-spatial-resolution, full vector field encoding, multiple MRE phases and high signal-to-noise-ratio (SNR), which usually results in long acquisition time. In this regards, spiral acquisition has been developed in spin echo sequence with multislice/multislab schemes4-5, providing improved tradeoff between resolution, SNR and scan time. Nevertheless, spiral acquisition in gradient echo (GRE) MRE has not been fully developed due to lower SNR. In this study, a novel 3D multishot, variable density spiral staircase (SSC) acquisition is proposed for GRE-MRE. We achieve high resolution by using improved through-plane parallel imaging inherent from the proposed SSC trajectory with much less SNR penalty and further boost the displacement-SNR by exploiting a MEG of 2 wave-period duration. In vivo human experiments have demonstrated that the proposed method could provide high-SNR shear wave and stiffness maps comparable to a standard single-shot spin-echo echo planar imaging scan and high resolution (e.g., 1.8×1.8×1.8mm3) whole-brain MRE is possible within 5 minutes.METHODS
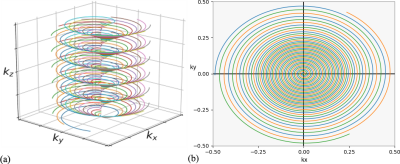
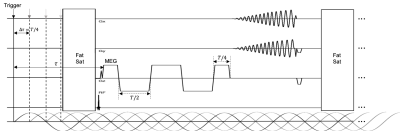
1) Data Acquisition: The proposed multishot, variable density 3D SSC trajectory is illustrated in Fig. 1. SSC uniformly distributes M groups of N spiral arms (each rotates by n*360°/N) along kz axis. Each set of M spiral arms (all have the same rotation about kz) is shifted by n*Δkz/N (Δkz is the Nyquist distance), resulting in a linear phase variation as a function of slice location. One advantage of this feature is that it introduces additional incoherence among image slices, leading to improved through-plane parallel imaging with much less SNR penalty.The pulse sequence for the proposed method is shown in Fig. 2. To enhance the displacement SNR, a motion encoding gradient (MEG) of a duration of 2 wave period (T) is adopted. The multishot, variable density spiral-out readout follows next, achieving a short TE of 35 ms approximately optimal for phase imaging in the brain at 3T6. Note that the proposed technique provides flexibility in trade-off among resolution, SNR, spatial-blurring and scan time. Given a desired resolution, acquisition parameters such as through/in-plane acceleration factor, spiral readout time, variable density can be adjusted accordingly to meet a feasible scan time with sufficient SNR.
2) Image Reconstruction: After obtaining the under sampled SSC data, spiral images from each MRE phase and MEG direction were reconstructed by solving the following optimization problem:
$$\hat{\rho}=\underset{\rho}{\operatorname{argmin}} \sum_{l=1}^{L}\left \| G_{xy}^{H}FP_zS_l\rho - d_l \right \|^2_2 + \lambda R(\rho)$$
where $$$\rho$$$ denotes the target image function, $$$S_l$$$ the $$$l$$$-th coil sensitivity profile, $$$P_z$$$ the linear phase compensation term for SSC along z, $$$F$$$ the Fourier transform operator, $$$G_{xy}$$$ the 2D gridding operator and $$$H$$$ the Hermitian conjugate operator. $$$d_l$$$ contains the spiral data from the $$$l$$$-th coil. $$$R(·)$$$ is a regularization function (e.g., Tikhonov regularization) which allows to incorporate additional prior information. After reconstruction, spiral images were processed using a conjugate gradient deblurring method7 with a known $$$B_0$$$ map to correct the off-resonance induced blurring. Both the sensitivity and $$$B_0$$$ maps were obtained from a low-resolution prescan.
3) MRE inversion: Given the deblurred images, the motion phase component along each axis can be calculated according to the motion encoding directions. With the curl of the motion phase determined8, the shear modulus is reconstructed via direct inversion of the Helmholtz equation9.
RESULTS
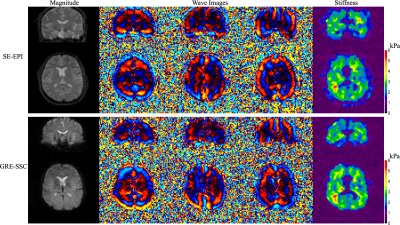
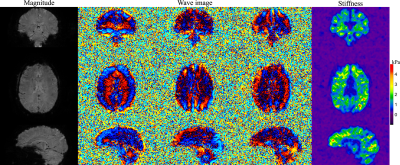
In vivo experiments were performed on a 3T Phillips system (Ingenia Elition X) with a 14-channel receiver head coil on two healthy subjects in accordance with the institutional IRB and written informed consent. Brain MRE data at two different spatial resolutions were acquired using the proposed method (FOV 240×240×120 mm3, TR/TE 67/35 ms, FA 23°, 2× undersampling along kz, wave frequency 60Hz, 4 MRE phases, 6 MEG directions) in 1.5 and 4.5 minutes respectively ( 16 ms spiral-readout, 2 spiral-shots, 1.4× slice oversampling for 3×3×3 mm3 and 18.6 ms spiral-readout, 4 spiral-shots, 1.2× slice oversampling for 1.8×1.8×1.8 mm3). To validate the stiffness images from the proposed method, a standard single-shot 2D spin-echo EPI MRE sequence was also conducted at 3×3×3 mm3 resolution for comparison (TR/TE 4000/67ms, FA 90°, scan time 96 seconds).MRE results generated from both methods were compared in Fig. 3. The proposed 3D gradient-echo acquisition provides high-SNR shear wave and stiffness images comparable to those from the 2D SE-EPI scan, while the conventional 2D scan will suffer from phase jitters across slices. The high-resolution results from another subject using the proposed method are shown in Fig. 4 with clearly identified brain regions (e.g., ventricles) in the stiffness maps, demonstrating the capability of the proposed method for high-quality MRE of the whole-brain.
CONCLUSION
A novel 3D GRE multi shot, variable density spiral staircase acquisition is proposed for whole-brain MRE. This technique integrates a 3D SSC trajectory, parallel imaging and a new MEG to achieve fast and high quality in vivo mapping of tissue mechanical property in the brain. Future work may include further optimizing scan parameters to provide better trade-off between SNR, resolution and scan time.Acknowledgements
This work is supported in part by a research funding from Philips.References
1. Murphy MC, Huston J, Jack CR, Glaser KJ, Manduca A, Felmlee JP, Ehman RL. Decreased brain stiffness in Alzheimer’s disease determined by magnetic resonance elastography. J Magn Reson Imaging 2011;34:494–498.
2. Streitberger K-J, Sack I, Krefting D, Pfuller C, Braun J, Paul F, Wuerfel J. Brain viscoelasticity alteration in chronic-progressive multiple sclerosis. PLoS One 2012;7:e29888.
3. Freimann FB, Streitberger K-J, Klatt D, Lin K, McLaughlin JR, Braun J, Sprung C, Sack I. Alteration of brain viscoelasticity after shunt treatment in normal pressure hydrocephalus. Neuroradiology 2012; 54:189–196.
4. Johnson CL, McGarry MD, Van Houten EE, et al. Magnetic resonance elastography of the brain using multishot spiral readouts with self-navigated motion correction. Magn Reson Med. 2013;70(2):404–412
5. Johnson CL, Holtrop JL, McGarry MD, et al. 3D multislab, multishot acquisition for fast, whole-brain MR elastography with high signal-to-noise efficiency. Magn Reson Med. 2014;71(2):477–485
6. Wu B, Li W, Avram AV, Gho SM, Liu C. Fast and tissue‐optimized mapping of magnetic susceptibility and T2* with multiecho and multi‐shot spirals. NeuroImage 2012;59:297–305.
7. Wang D, Zwart NR, Pipe JG. Joint water–fat separation and deblurring for spiral imaging. Magn Reson Med. 2018;79(6):3218–3228
8. Glaser KJ, Ehman R. MR Elastography Inversions Without Phase Unwrapping. ISMRM, 2009.
9. Manduca A, Oliphant TE, Dresner MA, Mahowald JL, Kruse SA et al. Magnetic resonance elastography: Non-invasive mapping of tissue elasticity. Med Image Anal 2001; 5: 237-254
Figures