0081
Deep learning-based sCTs with uncertainty estimation from heterogeneous paediatric brain MRI
Matteo Maspero1,2, Laura G Bentvelzen1,2, Mark H F Savenije1,2, Enrica Seravalli1, Geert O R Janssens1,3, Cornelis A T van den Berg1,2, and Marielle E P Philippens1
1Radiotherapy, UMC Utrecht, Utrecht, Netherlands, 2Computational imaging group for MR diagnostic & therapy, UMC Utrecht, Utrecht, Netherlands, 3Paediatric Oncology, Princess Maxima Center, Utrecht, Netherlands
1Radiotherapy, UMC Utrecht, Utrecht, Netherlands, 2Computational imaging group for MR diagnostic & therapy, UMC Utrecht, Utrecht, Netherlands, 3Paediatric Oncology, Princess Maxima Center, Utrecht, Netherlands
Synopsis
The feasibility of radiotherapy dose calculations for brain tumours from MRI acquired with a heterogeneous set of acquisition protocols on paediatric patients was investigated using a combination of networks trained on orthogonal planes to estimate the uncertainty of the generated sCT.
Background & objectives
To enable accurate MR-only dose planning, synthetic-CTs (sCTs) need to be generated1,2. Recently, convolutional networks have been proposed as a general-purpose solution to image-to-image translation problems enabling MRI-based dose calculations3,4.sCTs used in current clinical practice are generated by fixed MRI protocols, which restricts developments on MRI protocols and cross-platform usage. To date, it is unclear how a network would perform with variable imaging protocols, and, additionally, how the quality of sCT can be verified in daily clinical practice.
This work aims at assessing: (1) the feasibility of dose calculations from MRI acquired with a heterogeneous set of acquisition protocols; (2) investigating whether using different orthogonal planes to train a network to generate sCT can be beneficial; (3) proposing a combination of networks trained on multiple orthogonal planes to estimate the uncertainty of the generated sCT.
Material & methods
All paediatric patients (pts) undergoing radiotherapy for primary brain tumours between July 2015 and May 2019 were considered in this study. Patients underwent a planning CT in the treatment position at the radiotherapy department (Brilliance Big Bore, Philips Healthcare, Ohio, USA), while MRI could have been acquired in other departments or centres. Inclusion criterion was the presence of a 3D T1w spoiled gradient-recalled echo (SPGR) and a planning CT. All pts were included with imaging protocols varying among pts, as shown in Table 1.A conditional generative adversarial network (cGAN)5 was used to learn paired mapping from MRI to CT. Train/validation/test sets were created maintaining the heterogeneity of pts’ demographic in the three sets. The validation set was used to aid hyperparameter optimisation and find at which epoch perform early-stopping. CT images were rigidly registered and resampled to the 3D T1w SPGR images. Three cGANs were trained separately on 30 patients in the three orthogonal planes for 100 epochs (18-36 hrs on a GPU, NVIDIA). The networks were validated on 10 pts performing early-stopping. Final models were trained for 30 epochs on 40 pts and tested on the remaining 12 pts producing three sCTs per patient. For each patient, mean (±1σ) of the three views were calculated for each voxel obtaining a combined sCT (CsCT) and an uncertainty map, respectively. For the pts in the test set, the CsCT and sCTs obtained training in each orthogonal plane were evaluated in terms of mean absolute error (MAE) and dice coefficient of the bony structure (DiceBone) against the planning CT. Also, non-parametric Wilcoxon signed-rank tests were conducted. Dose recalculation of clinical plans was performed on 10 test pts on CsCT in the Monaco treatment planning system (Elekta AB, Sweden) on a 2 mm3 grid. Dose distributions were analysed through voxel-based dose differences, dose-volume histograms (DVH), and γ-analysis.
Results & discussion
In total, 52 patients, age=2-18 years-old, male/female ratio=25/27, and affected by diffuse astrocytic & oligodendroglial (n=18), embryonal (n=8), germ cell (n=6), sellar region (n=6), ependymal (n=5), or other rare tumours (n=9) have been included. Applying the trained cGANs to the three planes of a single patient (Figures 1-2) required about 20 s. An MAE of 75±15HU (mean±1σ, range:43-85) was obtained in the intersection of the body contours between CT and CsCT (Fig3). When comparing the results to an adult population of pts affected by brain tumours, one can observe that MAE is on average slightly higher but comparable to work presented in the literature3,6,7.The CsCT performed significantly better (p<0.001) than sCTs from networks trained in every single plane: sagittal and coronal planes had the lowest MAE (84±18 and 84±17, respectively), followed by transverse (89±19) plane. Significant differences were observed for MAE between the sCT trained on the transverse plane and the other two planes. Data unbalancing was observed in the number of slices used for training: 12056, 13328, 8573 images were used for training in the transverse, coronal, sagittal plane, respectively. This may contribute to the different performance among the models.
Current investigations are verifying the consistency of the results depending on the patients’ age, sex, tumour type, tumour location, magnetic field strength, FOV and scan with/without gadolinium. So far, we noticed a lower CsCT image quality for young patients (<3 years-old) scanned with gadolinium, as, for example in Figure 2. However, the dose accuracy was not hindered.
A dose difference of -0.1± 0.4% was obtained on the D>50% of the prescribed dose and mean γ-2%,2mm pass rate of 99.2±1.5% (Table 2), which can be considered as sufficiently accurate for clinical use. These results are in line with the previous works3,6,7. Uncertainty maps were qualitatively reviewed for each patient noticing high values at the body contours and air cavities. We are currently searching for quantitative metrics on this uncertainty map that could be used to assess the sCT quality in the absence of CT.
To our knowledge, this is the first time that sCTs are generated using convolutional neural networks for a paediatric population.
Conclusion
Accurate MRI-based dose calculation using a combination of three orthogonal planes for sCT generation is feasible for paediatric brain cancer patients, even when training on a heterogeneous dataset. By combining the different planes an uncertainty map was estimated, which enabled assessing the sCT quality.Acknowledgements
We gratefully acknowledge the support of NVIDIA Corporation with the donation of the Titan Xp GPU used for prototyping this research.References
- Edmund JM, Nyholm T. A review of substitute CT generation for MRI-only radiation therapy. Radiat Oncol. 2017;12(1):28.
- Kerkmeijer LG, Maspero M, Meijer GJ, van Zyp JV, de Boer HC, van den Berg CA. Magnetic resonance imaging only workflow for radiotherapy simulation and planning in prostate cancer. Clinic Oncol. 2018;30(11):692-701.
- Maspero M, Savenije MH, Dinkla AM, Seevinck PR, Intven MP, Jurgenliemk-Schulz IM, Kerkmeijer LG, van den Berg CA. Dose evaluation of fast synthetic-CT generation using a generative adversarial network for general pelvis MR-only radiotherapy. Phys Med Biol. 2018;63(18):185001.
- Dinkla AM, Wolterink JM, Maspero M, Savenije MH, Verhoeff JJ, Seravalli E, Išgum I, Seevinck PR, van den Berg CA. MR-only brain radiation therapy: Dosimetric evaluation of synthetic CTs generated by a dilated convolutional neural network. Int J Radiat Oncol Biol Phys. 2018;102(4):801-12.
- Isola P, Zhu JY, Zhou T, Efros AA. Image-to-Image Translation with Conditional Adversarial Networks. In2017 IEEE CVPR 2017:5967-5976).
- Kazemifar S, McGuire S, Timmerman R, Wardak Z, Nguyen D, Park Y, Jiang S, Owrangi A. MRI-only brain radiotherapy: Assessing the dosimetric accuracy of synthetic CT images generated using a deep learning approach. Radiotherapy and Oncology. 2019 Jul 1;136:56-63.
- Spadea MF, Pileggi G, Zaffino P, Salome P, Catana C, Izquierdo-Garcia D, Amato F, Seco J. Deep Convolution Neural Network (DCNN) Multiplane Approach to Synthetic CT Generation From MR images-Application in Brain Proton Therapy. International Journal of Radiation Oncology* Biology* Physics. 2019 Nov 1;105(3):495-503.
Figures
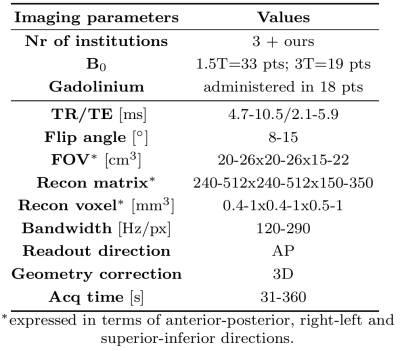
Table 1 MRI protocols of the 3D T1w spoiled gradient-recalled echo sequence used for sCT generation reporting the (min-max) of parameters values.

Figure 1 Example of MRI @3T, CsCT, CT, uncertainty map and DVH for a paediatric patient of 17 years old affected by glioblastoma. This patient had MAE=45 HU, DiceBone=0.87 for the combined sCT.
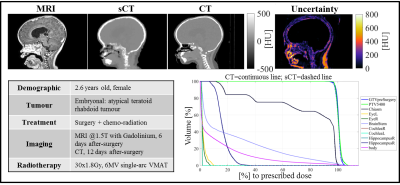
Figure 2 Example of MRI @1.5T with the administration of gadolinium, CsCT, CT, uncertainty map and DVH for a paediatric patient affected by an atypical teratoid rhabdoid tumour. This patient had MAE=85 HU, DiceBone=0.81 for the combined sCT.
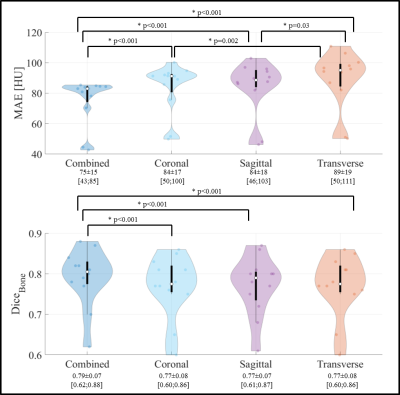
Figure 3 Violin plots of mean absolute error (MAE) in the body contour and dice coefficients of bony structures (DiceBone) of sCTs against CT. The p-value <0.05 of the Wilcoxon signed-rank test are reported.
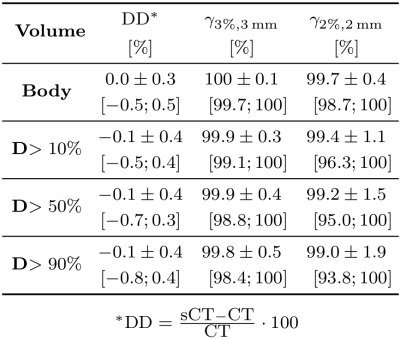
Table 2 Dose comparison between CsCT and CT calculated as mean (±1σ) and range ([min; max]) of relative dose difference (DD) and γ pass rate on several dose threshold levels.