4774
DCTV-Net: Model based Convolutional Neural Network for dynamic MRI1Shenzhen Institutes of Advanced Technologies, Xili Nanshan, China, 2Department of Biomedical Engineering and Department of Electrical Engineering, The State University of New York, Buffalo, NY, United States
Synopsis
Compressive sensing MRI (CS-MRI) is a popular technique to accelerate MR dynamic imaging. Nevertheless, the reconstruction is normally time-consuming and its parameters have to be hand-tuned To address this challenge, we solve a CS-based dynamic MR imaging problem by adopting the Alternating Direction Method of Multipliers (ADMM) iteration method with the most popular deep learning technique. Specifically, we introduce a deep network structure, dubbed as DCTV-NET, for dynamic magnetic resonance image reconstruction from highly under-sampled k-t space data. Experimental results demonstrate that our method is superior to the state-of-the-art dynamic MRI methods.
Introduction
Dynamic MR imaging is an important imaging tool for many clinical applications. However, it is fundamentally challenging to achieve fast imaging speed without sacrificing high spatial and temporal resolutions. Compressive sensing MRI (CS-MRI) is a popular technique to accelerate MR dynamic imaging, such as k-t SPARSE, k-t PCA and so on so forth 2-4. Nevertheless, the reconstruction is normally time-consuming and its parameters have to be hand-tuned. To address this challenge, we solve a CS-based dynamic MR imaging problem by adopting the Alternating Direction Method of Multipliers (ADMM) iteration method6 with the most popular deep learning technique7-14. Specifically, we proposed a deep network structure, dubbed as DCTV-NET, which expands the iterative ADMM process to learn a convolutional neural network. In the convolution layer, we use DCT in the spatial domain and TV in the temporal to explore the redundancy and improve the reconstruction accuracy. Experiments demonstrate that the proposed scheme can effectively reconstruct dynamic MR images with higher accuracy in shorter time.Theory and method
The essence of the proposed approach is to integrate the merits of model based method in finding theoretically optimal or sub-optimal solutions and strengths of deep learning based methods in automatically learning the weighting parameters with higher reconstruction speed. The dynamic MR imaging reconstruction problem can be described as follows
$$\min_{x,z}\frac{1}{2}\Vert Ax-y\Vert_2^2+\sum_{l=1}^L\lambda_lg(D_lz) \ \ s.t.\ z=x$$
where A=PF is a measurement matrix with P as the undersampling pattern and F as the Fourier transform; 𝑥 is an image to be reconstructed; y is the under-sampled k-space data. $$$\lambda _l$$$ is a regularization parameter. $$$g(\cdot)$$$ is a regularization function related to data prior. $$$D_l$$$ is a filtering operation. z is the auxiliary variable in the spatial domain. Its augmented Lagrangian equation could be described as follows:
$$\mathcal L_p(x,z,\alpha)=\frac{1}{2}\Vert Ax-y\Vert_2^2+\sum_{l=1}^L\lambda_lg(D_lz)+\langle \alpha,z-x\rangle+\frac{\rho}{2}\Vert z-x\Vert_2^2$$
Then, it could be transformed into the following sub-problems:
$$\begin{cases}\arg\min\limits_x \frac{1}{2}\Vert Ax-y \Vert_2^2 + \langle \alpha,z-x \rangle+\frac{\rho}{2}\Vert z-x\Vert_2^2 \\ \arg\min\limits_z \sum_{l=1}\limits^L \lambda_lg(D_lz)- \langle \alpha,z-x \rangle +\frac{\rho}{2}\Vert z-x\Vert_2^2 \\arg\min\limits_a\langle \alpha,z-x \rangle\end{cases}$$
The sub-problems have the following solutions, where the auxiliary variable z employs a gradient-descent algorithm. $$$\beta=\alpha /\rho$$$ is the scaled multiplier for Lagrangian. $$$\widetilde{\eta}$$$ is an update rate.
$$\begin{cases} x^{(n)}=F^T\left(P^TP+\rho^{(n)}I\right)^{-1}\left[P^Ty+\rho^{(n)}F\left(z^{(z-1)}-\beta^{(n-1)}\right)\right]\\z^{(n,k)}= \mu_1z^{(n,k-1)}+\mu_2\left(x^{(n)}+\beta^{(n-1)}\right) - \sum\limits_{l=1}^L \widetilde{\lambda}_l D_l^T \mathcal H\left(D_l z^{(n,k-1)}\right) \\\beta^{(n)}=\beta^{(n-1)}+\widetilde{\eta}\left(x^{(n)}-z^{(n)}\right) \end{cases}$$
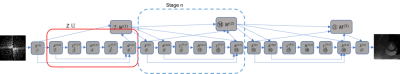
Our defined network, DCTV-NET, is shown in Fig.1. X is the reconstruction layer $$$(X^{(n)})$$$. Z is the denoising layer and is decomposed into an additional layer $$$(A^{(n,k)})$$$, convolution layers $$$(C_1^{(n,k)},C_2^{(n,k)})$$$ and a nonlinear transform layer $$$(H^{(n,k)})$$$. M is the multiplier update layer $$$(M^{(n)})$$$.
Experiment
We collected 65 fully sampled dynamic cardiac MR data using a 3T MRI system (SIEMENS MAGNETOM Trio) with a T1-weighted cine flash sequence. The scan parameters were TR/TE=2.58/49.14ms, number of slices=25, slice thickness=8mm, FOV=280mm. We cut data into $$$126\times126\times16$$$ volumes and perform Fourier transform to obtain the K-space data. Images with 5x,7x, 9x and 11x 1D random under-sampling were obtained by applying the corresponding masks. 60 data were used for network training, and 5 for network testing. The model training was implemented on an Intel Xeon (R) CPU E5-2640 V4 @2.40GHz × 40 with 64G memory.Results and discussion
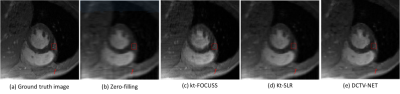
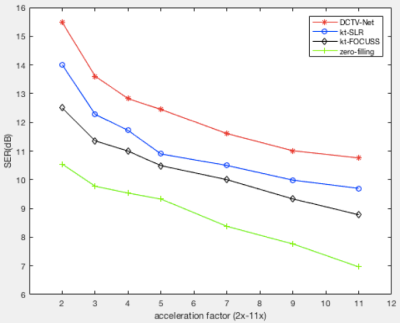
Fig.2 presents the reconstructions of the Zero-filling (b), kt-FOCUSS2 (c), kt-SLR7 (d) and the proposed method (e) retrospectively using 1D random undersampling mask at 5x acceleration. Table.1 shows the quantitative results. Both qualitative and quantitative results demonstrate that the proposed method is superior to the state-of-the-art method. In detail, PSNR value of the proposed method is nearly 4dB larger than Kt-SLR. Similarly, improvements in SSIM and test time are observed. Fig.3 compares the different algorithms' average signal to error ratio (SER) of five test data when acceleration factor varies from 2x to 11x. The figure shows very clearly that our results have smaller errors and higher reconstruction accuracy. Where $$$SER=-10\log_{10}\frac{\Vert \hat {x} (y,\Theta)-x^{gt} \Vert _F^2}{\Vert x^{gt} \Vert _F^2}$$$Conclusion
This work proposes a model based deep network DCTV-NET for the reconstruction of dynamic MR images. It not only possesses the merits of model based method in finding theoretically optimal or sub-optimal solutions, but also has strengths of deep learning based methods in automatically learning the weighting parameters with faster reconstruction speed. Experimental results show that our method could achieve improved result in much shorter time compared to other three methods.Acknowledgements
This work was supported by funding from the National Natural Science Foundation of China (61601450, 61871371, and 81830056), Science and Technology Planning Project of Guangdong Province (2017B020227012).References
1. Michael Lustig, David L Donoho, Juan M Santos, and John M Pauly. Compressed sensing mri. IEEE Journal of Signal Processing, 25(2):72–82, 2008.
2. H. Jung, K. Sung, K. S. Nayak, E. Y. Kim, and J. C. Ye, “k-t FOCUSS: A general compressed sensing framework for high resolution dynamic MRI,” Magnetic Resonance in Medicine, vol. 61, no. 1, pp. 103–116, 2009.
3. M. Lustig, J. Santos, D. Donoho, and J. Pauly, “k-t SPARSE: High frame rate dynamic MRI exploiting spatio-temporal sparsity,” in International Society on Magnetic Resonance in Medicine 2006.
4. F. Knoll, K. Bredies, T. Pock, and R. Stollberger, “Second order total generalized variation (TGV) for MRI,”Magnetic resonance in medicine, vol. 65, no. 2, pp. 480–491, 2011.
5. S. Poddar, Mathews Jacob, “Dynamic MRI using SmooThness Regularization on Manifolds (SToRM)”, IEEE, p1106-1115,2015
6. Y. Yang, J. Sun, H. Li, Zongben Xu, “ADMM-Net: A Deep Learning Approach for Compressive Sensing MRI”,2017
7. S. Lingala, Y. Hu, E. DiBella, and M. Jacob, “Accelerated dynamic MRI exploiting sparsity and low-rank structure: k-t SLR,” IEEE Trans. Med. Imag., vol. 30, no. 5, pp. 1042–1054, May 2011.
8. Han, Yoseob, Jaejun Yoo, Hak Hee Kim, Hee Jung Shin, Kyunghyun Sung, and Jong Chul Ye. "Deep learning with domain adaptation for accelerated projection‐reconstruction MR." Magnetic resonance in medicine 80, no. 3 (2018): 1189-1205.
9. J. Schlemper, J. Caballero, J.V. Hajnal, A. Price, D. Rueckert, “A Deep Cascade of Convolutional Neural Networks for Dynamic MR Image Reconstruction”, IEEE TMI, DOI: 10. 1109/TMI.2017.2760978 (2017)
10. Qin, Chen, Joseph V. Hajnal, Daniel Rueckert, Jo Schlemper, Jose Caballero, and Anthony N. Price. "Convolutional recurrent neural networks for dynamic MR image reconstruction." IEEE transactions on medical imaging (2018).
11. Zhu, Bo, Jeremiah Z. Liu, Stephen F. Cauley, Bruce R. Rosen, and Matthew S. Rosen. "Image reconstruction by domain-transform manifold learning." Nature 555, no. 7697 (2018): 487.
12. Eo, Taejoon, Yohan Jun, Taeseong Kim, Jinseong Jang, Ho‐Joon Lee, and Dosik Hwang. "KIKI‐net: cross‐domain convolutional neural networks for reconstructing undersampled magnetic resonance images." Magnetic resonance in medicine (2018).
13. K. Hammernik, T. Klatzer, E. Kobler, M. P. Recht, D. K.Sodickson, T. Pock, and F. Knoll, “Learning a variationalnetwork for reconstruction of accelerated MRI data,”Magnetic Resonance in Medicine, vol. 79, no. 6, pp.3055–3071, 2018.
14. H.K. Aggarwal, M.P Mani, and Mathews Jacob, MoDL: Model Based Deep Learning Architecture for Inverse Problems, IEEE Transactions on Medical Imaging, 2018
Figures