4710
Rapid Image Reconstruction of Single-Shot Coronary Quiescent-Interval Slice-Selective (QISS) MRA and Late Gadolinium-Enhanced MRI using Deep Learning1Biomedical Engineering, Northwestern University, Evanston, IL, United States, 2Radiology, Northwestern University, Chicago, IL, United States
Synopsis
While compressed sensing (CS) enables highly-accelerated cardiac MRI acquisitions, its lengthy image reconstruction may limit clinical translation. Deep learning (DL) is capable reconstructing undersampled images with clinically acceptable reconstruction times. The purpose of this study was to build, train, and validate a deep learning framework for rapidly reconstructing highly-accelerated cardiac MR images, where CS reconstructed images are used as reference.
Introduction
Compressed sensing (CS)1 is a proven framework for highly accelerating cardiac MRI acquisitions, but its lengthy image reconstruction may limit clinical translation. One approach to overcome this problem is to use a deep learning (DL) framework2-5 instead. In this study, we sought to build, train, and validate a DL network for rapidly reconstructing highly-accelerated cardiac MRI images. Specifically, we used raw data from previously described single-shot coronary Quiescent-Interval Slice-Selective (QISS) MRA6 and single-shot late gadolinium-enhanced (LGE)7 studies. The purpose of this study was to build and train a DL framework based on single-shot/two-shot QISS data sets and validate the trained network for both single-shot QISS images and single-shot LGE images with similar radial k-space sampling patterns.Methods
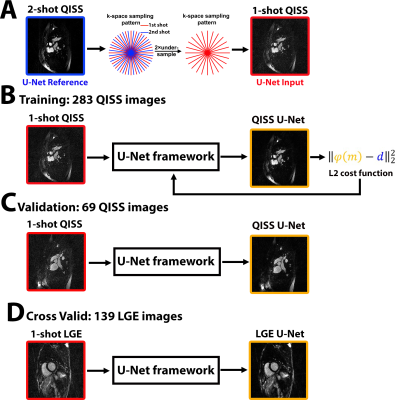
Human Subject & Pulse Sequence: We obtained two-shot coronary QISS MRA data from a prior study at 1.5T (Siemens Aera) with radial k-space sampling. Relevant imaging parameters include: FOV of 180 × 180 mm, matrix size of 128 × 128, slice thickness of 2.1 mm, 96 rays (48 rays per heart beat) with 13.123° increments, flip angle 140°, TE = 1.9 ms, TR = 736 ms, echo spacing (ESP) = 3.7 ms, inversion time (TI) = 650 ms. In total 26 pediatric patients (mean age = 16.4 ± 7.9 years; 16 boys and 10 girls) were included in this study. Single-shot, free-breathing LGE with radial k-space sampling was implemented on 1.5T (Avanto, Aera, Siemens) and 3T (Skyra, Siemens) scanners. Relevant imaging parameters include: FOV of 300 × 300 mm, matrix size of 256 × 256, slice thickness of 8 mm, 42 rays with 32.040° increments, flip angle 75°. We enrolled 9 patients (mean age = 57.6 ± 16.8 years; 3 males; 6 females) for this study.
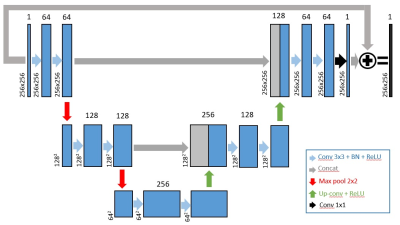
Image Reconstruction: Two-shot QISS data were retrospectively undersampled by a factor of two and reconstructed using CS, where total variation (TV) was used as the sparsifying transform. A deep convolutional neuron network (CNN) with 2D U-Net structure (layer depth = 3, 64 features on the first layer, batch size = 4, GPU (P100 Tesla, NVIDIA) based tensorflow framework in Python) was built as shown in Figure 1. For training, we fed zero-filled, 1-shot and 2-shot QISS images from first 20 patients (283 images) as input/output pairs to train the U-Net. For validation, zero-filled, 1-shot images and CS reconstructed images from the remaining 6 patients (69 images) were used as input/output pairs to the trained U-Net. For cross validation, we fed zero-filled, single-shot LGE images and CS reconstructed images from 9 patients (139 images) were used as input/output pairs.
Quantitative Metrics: Using single-shot, CS reconstructed images (for both QISS and LGE) as reference, we calculated the structural similarity index (SSIM), Dice similarity coefficient (DSC), and normalized root mean square error (NRMSE) to infer data fidelity. A mask was used to exclude signal-free pixels in the background.
Results
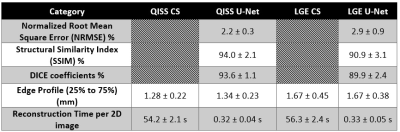
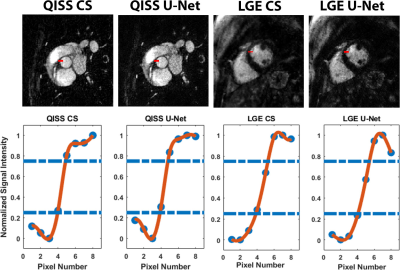
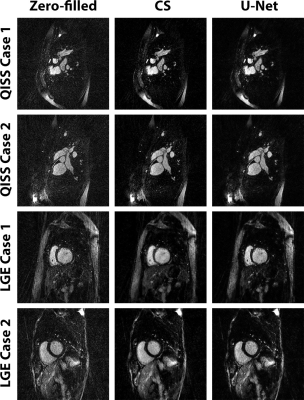
Figure 4 shows representative QISS MRA images obtained with CS and CNN reconstruction frameworks for three different patients. For QISS MRA, as summarized in Table 1, compared with CS, CNN produced high data fidelity (DSC=93.6 ± 1.1%; SSIM=94.0 ± 2.1%, NRMSE=2.2 ± 0.3%). The edge sharpness (1.28 ± 0.22 mm for CS vs. 1.34 ± 0.23 mm for CNN) was not significantly different (P > 0.38). The mean reconstruction time of CNN (0.32 s) was 99% shorter than CS (54s). For LGE, compared with CS, CNN produced high data fidelity (DSC=89.9 ± 2.4%; SSIM=90.9 ± 3.1%, NRMSE=2.9 ± 0.9%). The edge sharpness (1.67 ± 0.45 mm for CS vs. 1.67 ± 0.38 mm for CNN) was not significantly different (P > 0.98).Discussions
This study demonstrates that a U-Net framework reconstructs accelerated QISS MRA and LGE images with high data fidelity at 99% shorter reconstruction time than CS, thereby making inline reconstruction possible. A future study is warranted to investigate additional approaches to increasing data fidelity by incorporating a fidelity term.Acknowledgements
This work was supported in part by the following grants: NIH R01HL116895, R01HL138578, R21EB024315, R21AG055954References
1. Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med 2007;58(6):1182-1195.
2. Kerstin H, Teresa K, Erich K, P. RM, K. SD, Thomas P, Florian K. Learning a variational network for reconstruction of accelerated MRI data. Magnetic Resonance in Medicine 2018;79(6):3055-3071.
3. Han Y, Yoo J, Kim HH, Shin HJ, Sung K, Ye JC. Deep learning with domain adaptation for accelerated projection-reconstruction MR. Magn Reson Med 2018;80(3):1189-1205.
4. Wang S, Su Z, Ying L, Peng X, Zhu S, Liang F, Feng D, Liang D. Accelerating magnetic resonance imaging via deep learning. 2016 13-16 April 2016. p 514-517.
5. Schlemper J, Caballero J, Hajnal JV, Price AN, Rueckert D. A Deep Cascade of Convolutional Neural Networks for Dynamic MR Image Reconstruction. IEEE Transactions on Medical Imaging 2018;37(2):491-503.
6. Shen D, Edelman RR, Robinson JD, Haji-Valizadeh H, Messina M, Giri S, Koktzoglou I, Rigsby CK, Kim D. Single-Shot Coronary Quiescent-Interval Slice-Selective Magnetic Resonance Angiography Using Compressed Sensing: A Feasibility Study in Patients With Congenital Heart Disease. J Comput Assist Tomogr 2018.
7. Haji-Valizadeh H, Collins J, Lee D, Carr J, Kim D. High resolution, single-shot LGE MRI with compressed sensing and radial k-space sampling. In: Proceedings of the 25rd Annual Meeting of ISMRM, Honolulu, HI 2017. Abstract No. 3255.
Figures