3714
Quantifying and Correcting for the Assumptions of the Static Dephasing Regime in Quantitative BOLD1Institute of Biomedical Engineering, Department of Engineering Science, University of Oxford, Oxford, United Kingdom, 2Wellcome Centre for Integrative Neuroimaging, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom, 3School of Life Sciences, University of Nottingham, Nottingham, United Kingdom
Synopsis
The effect of diffusion is believed to cause underestimation of oxygen extraction fraction (OEF) in streamlined quantitative BOLD (qBOLD). We propose physically motivated modifications to the qBOLD model in order to account for this effect, and demonstrate, using Monte Carlo simulations, and analysis of healthy in vivo data, that simple scalar corrections to the inferred reversible relaxation rate $$$R_2'$$$, can substantially reduce the error in OEF and deoxygenated blood volume (DBV) estimation. These corrections have the potential to be improved further by taking other physiological and acquisition-related parameters into account.
Introduction
Quantitative BOLD (qBOLD) is used to quantify oxygen extraction fraction (OEF) and deoxygenated blood volume (DBV) in the brain. It models changes in the measured reversible relaxation rate $$$R_2'$$$ as a function of OEF and DBV, under the assumption that diffusion does not have an appreciable effect (the static dephasing regime – SDR)[1]. However, Monte Carlo simulations have shown that diffusion has a significant effect on the measured signal[2,3]. Efforts to incorporate diffusion into the qBOLD model[4] have generally focused on deriving new models from first principles[5], or on phenomenologically explaining simulated data[6]. We present physically motivated modifications to the asymptotic solutions of the SDR model (which have been shown to be robust over a range of parameter values[7]) that enable accurate parametrization of simulated and in vivo data.Theory
The streamlined qBOLD method uses asymmetric spin echo (ASE) to quantify $$$R_2'$$$ and DBV, from which OEF can be computed[8,9]. In the regime of large spin-echo offsets $$$\tau$$$, the ASE signal is: $$$\operatorname{ln}S(\tau)=-R_2'\cdot{}\tau$$$, where
$$R_2'=k\cdot{}Hct\cdot{}OEF\cdot{}DBV\tag{1}$$
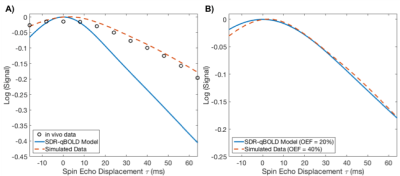
k is a constant that subsumes vessel geometry, susceptibility differences, and magnetic field strength, and Hct is the fractional haematocrit (assumed constant). When applied to real data, and compared to Monte Carlo simulations, this method underestimates $$$R_2'$$$ (hence OEF), as illustrated in Figure 1. This may be the result of diffusive dephasing[3]. To rectify this, we propose defining $$$R_2'$$$:
$$R_2'=\kappa\cdot{}[dHb]^\beta\cdot{}DBV\tag{2}$$
where [dHb] is the deoxyhaemoglobin concentration ($$$[dHb]=Hct\cdot{}OEF/k'$$$) which is related to the BOLD signal by exponent $$$\beta$$$[10,11]; and $$$\kappa$$$ is a scaling constant that subsumes k and k'. We expect that $$$\kappa$$$ will depend on OEF, and define it parametrically:$$\kappa=A+B\cdot{}OEF\tag{3}$$where A and B will be defined by optimization. We do not expect such variance in $$$\beta$$$.
Methods
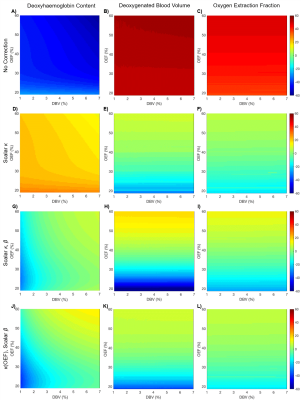
Simulations: In MATLAB (MathWorks, Natick, MA), Monte Carlo methods were used to generate simulated ASE signals for a 100x100 matrix of OEF and DBV in physiological ranges[3]. This involved filling a space with randomly oriented cylinders following a known distribution of vessel radii[12] and calculating the phase accrued by protons that diffuse around them. Three optimizations were performed; (i) minimization of the error in OEF estimate using $$$\kappa$$$ alone, (ii) minimization of error using $$$\kappa$$$ and $$$\beta$$$ and (iii) minimization of error where $$$\kappa$$$ is a function of OEF and $$$\beta$$$ is scalar.
Data Acquisition: Five healthy participants (24-35, 2F) were scanned using a 3T Verio system (Siemens Healthineers, Germany). GESEPI-ASE scans[8] were acquired (FOV=240mm2, 96x96 matrix, eight 5mm slices, TR/TE=3s,82ms, TIFLAIR=1210ms, $$$\tau$$$=-16:64ms in steps of 8ms) along with a high-resolution anatomical for segmentation.
In Vivo Data Analysis: Variational Bayesian inference was performed using FABBER[13] to estimate $$$R_2'$$$ and DBV[9] using Equations 1-3 with optimized values. OEF was calculated voxel-wise from $$$R_2'$$$ and DBV, and average grey-matter values were obtained using anatomical data segmented with FSL-FAST[14].
Results
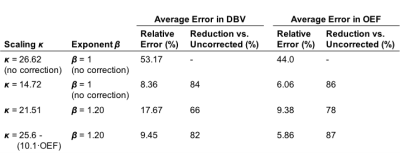
Simulations: Figure 2 shows the results of parameter estimates obtained using simulated data. Although the error in deoxyhaemoglobin content estimation is not uniform across OEF-DBV space, a value of $$$\kappa=14.72$$$ was found which minimized the average error, reducing it from 45.0% to 19.8%. Simultaneously optimizing $$$\kappa$$$ and $$$\beta$$$ ($$$\kappa=21.51$$$, $$$\beta=1.2$$$) reduced this error even further, to 6.9%. Applying scalar $$$\kappa$$$ correction to OEF and DBV estimation also reduced their relative errors, as shown in Table 1. The effects of including $$$\beta$$$ correction and of parametrically defining $$$\kappa$$$ as a function of OEF, are summarized in Table 1.
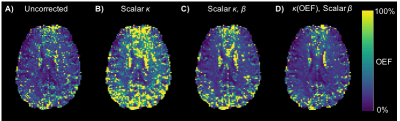
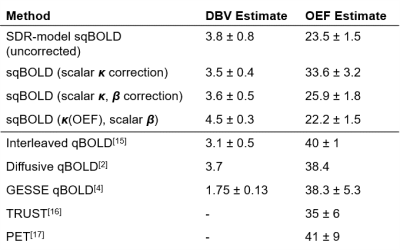
In Vivo Data: Figure 3 shows example OEF maps obtained from the several correction methods. Mean grey matter estimates of OEF and DBV were obtained using each, the group averages are shown in Table 2. Subject-wise ANOVA showed statistically significant differences between OEF distributions of all methods.
Discussion
Results from simulations and in vivo data show that using $$$\kappa$$$ scaling significantly increases the accuracy of single-parameter OEF and DBV estimation. Table 2 compares in vivo measurements produced by the above corrections, with those obtained from other methods. Correction using scalar $$$\kappa$$$ brings ASE-qBOLD OEF estimates closest to the literature values. The inclusion of $$$\beta$$$ exponent to [dHb], alongside parametric $$$\kappa$$$, did not improve estimation. It is likely that other factors besides OEF affect $$$\kappa$$$, such as DBV and TE, whose inclusion may improve estimation further.Conclusion
We propose that physically motivated modifications to the SDR qBOLD model can substantially improve its accuracy in estimating OEF and DBV. We demonstrate this using simulated data by substantially reducing the systematic error in estimates, and in vivo by producing estimates that are closer to expected literature values[2,4,15-17]. This approach can be extended to consider other confounds.Acknowledgements
This work was supported by funding from the Engineering and Physical Sciences Research Council (EPSRC) [grant number EP/K025716/1] and joint funding from the EPSRC and Medical Research Council (MRC) [grant number EP/L016052/1]. The Wellcome Centre for Integrative Neuroimaging is supported by core funding from the Wellcome Trust (203139/Z/16/Z).References
1. Yablonskiy DA, Haacke EM (1994). Theory of NMR Signal Behavior in Magnetically Inhomogeneous Tissues: The Static Dephasing Regime. Magn. Reson. Med., 32(6), 749–763.
2. Dickson JD, et al. (2010). Quantitative BOLD: the effect of diffusion. Journal of Magnetic Resonance Imaging, 32(4), 953–961.
3. Blockley NP, Holland NC, Stone AJ. (2016). The effect of diffusion on quantitative BOLD parameter estimates acquired with the Asymmetric Spin Echo technique. Proc ISMRM 2016, #3731.
4. He X, Yablonskiy DA (2007). Quantitative BOLD: Mapping of human cerebral deoxygenated blood volume and oxygen extraction fraction: Default state. Magn. Reson. Med., 57(1), 115–26.
5. Kiselev VG, Posse S (1999). Analytical Model of Susceptibility-Induced MR Signal Dephasing: Effect of Diffusion in a Microvascular Network. Magn. Reson. Med., 41, 499–509.
6. Dickson JD, et al. (2011). Quantitative phenomenological model of the BOLD contrast mechanism. Journal of Magnetic Resonance Imaging, 212, 17–25.
7. Domsch S, et al. (2018). Oxygen Extraction Fraction Mapping at 3 Tesla Using an Artificial Neural Network: A Feasibility Study. Magn. Reson. Med., 79(2), 890–899.
8. Stone AJ, Blockley NP. (2017). A streamlined acquisition for mapping baseline brain oxygenation using quantitative BOLD. Neuroimage, 147, 79–88.
9. Cherukara MT, et al. (2017). Bayesian Inference of Brain Oxygenation and Deoxygenated Blood Volume in Acute Stroke using Streamlined Quantitative BOLD. Proc. ISMRM 2017, #5269.
10. Ogawa S, et al. (1993). Functional brain mapping by blood oxygenation level-dependent contrast magnetic resonance imaging. A comparison of signal characteristics with a biophysical model. Biophysical Journal, 64(3), 803–812.
11. Blockley NP, Griffeth VEM, Buxton RB (2012). A general analysis of calibrated BOLD methodology for measuring CMRO 2 responses: Comparison of a new approach with existing methods. NeuroImage, 60, 279–289.
12. Sharan M, et al. (1989). A Compartmental Model for Oxygen Transport in Brain Microcirculation. Annals of Biomedical Engineering, 17(1), 13–38.
13. Chappell MA, et al. Variational Bayesian Inference for a Nonlinear Forward Model. IEEE Trans. Signal Process. 2009; 57(1):223–36.
14. Zhang Y, Brady MJ, Smith SM (2001). Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imag. 20(1), 45-57.
15. Lee H, Englund EK, Wehrli FW (2018). Interleaved quantitative BOLD: Combining extravascular R2’- and intravascular R2- measurements for estimation of deoxygenated blood volume and hemoglobin oxygen saturation. NeuroImage, 174, 420-431.
16. Lu H, Ge Y (2008). Quantitative evaluation of oxygenation in venous vessels using T2-relaxation-under-spin-tagging MRI. Magn. Reson. Med., 60, 357-363.
17. Derdeyn CP, et al. (2001). Comparison of PET Oxygen Extraction Fraction Methods for the Prediction of Stroke Risk. Journal of Nuclear Medicine, 42, 1195-1197.
Figures