3573
Biases of microstructure models in baby diffusion MRI1Biomedical Engineering Department, The University of North Carolina at Chapel Hill, Chapel Hill, NC, United States, 2Department of Radiology and BRIC, The University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
Synopsis
In this abstract, we show that some commonly used microstructure models do not perform as aspected when dealing with baby diffusion MRI, resulting in biased measurements. We found that this is mainly due to the greater water content in the developing brain. We show that the recently introduced Spherical Mean Spectrum Imaging (SMSI) [4] gives stable measurements in the presence of free water, making it well-suited for baby diffusion MRI.
Introduction
Probing brain microstructure with diffusion MRI provides crucial insights into the growth mechanisms of the developing brain, especially in the first years of life. However, baby diffusion MRI poses additional challenges owing to unmyelinated axons. The validity of many microstructure models have not be evaluated for baby diffusion MRI data. In this work, using synthetic data and longitudinal in-vivo diffusion MRI data, we assess 4 microstructure models for potential biases in relation to water content in the baby brain: Spherical Mean Technique (SMT), Multi-compartment Spherical Mean Technique (MC-SMT), Neurite Orientation Dispersion and Density Imaging (NODDI), and Spherical Mean Spectrum Imaging (SMSI).
Methods
We investigated four microstructure models in diffusion MRI including: Spherical Mean Technique (SMT) [1], Multi-compartment Spherical Mean Technique (MC-SMT) [2], Accelerated Microstructural Imaging via Convex Optimization (AMICO) of Neurite Orientation Dispersion and Density Imaging (NODDI) [3], and Spherical Mean Spectrum Imaging (SMSI) [4] using both synthetic and in-vivo data.
Synthetic data: We used the 6-shell Baby Connectome Project (BCP) gradient scheme to generate the data [5]. The normalized signal is constructed as: $$S = v_{iso}S_{iso} + (1-v_{iso})(v_{ec}S_{ec} + v_{ic}S_{ic})$$ Where $$$v_{\cdot}$$$ and $$$S_{\cdot}$$$ denote the volume fraction and normalized signal of each of the compartments: isotropic (iso), extra-cellular (ec), and intra-cellular (ic). We set $$$v_{iso}$$$ from 0 to 0.9, step size = 0.1, $$$v_{ec}=v_{ic}=0.5$$$, $$$\lambda_{\parallel}^{iso}=\lambda_{\bot}^{iso}=3 \times 10^{-3} mm/s^2$$$, $$$\lambda_{\parallel}^{ec}=\lambda_{\parallel}^{ic}=1.7 \times 10^{-3} mm/s^2$$$, $$$\lambda_{\bot}^{ec}=0.4 \times 10^{-3} mm/s^2$$$, and $$$\lambda_{\bot}^{ic}=0 \times 10^{-3} mm/s^2$$$. Rician noise was also added with a SNR of 30.
In-vivo data: We use longitudinal data of a baby subject collected at 54, 146, and 223 days after birth from the BCP data.
Results and Discussions:
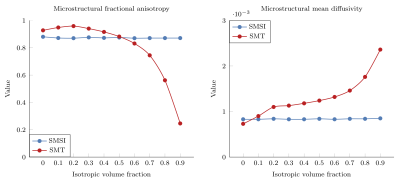
Microstructural Fractional Anisotropy ($$$\mu$$$FA) and Mean Diffusivity ($$$\mu$$$MD): Fig. 1 shows the micro-structural $$$\mu$$$FA and $$$\mu$$$MD from SMT and the free-water-eliminated $$$\mu$$$FA and $$$\mu$$$MD from SMSI with respect to the isotropic compartment volume fraction (IVF). SMSI results stay close to the ground truth (FA=$$$0.87$$$, MD=$$$0.7 \times 10^{-3}$$$) whereas SMT overestimates $$$\mu$$$FA when the IVF is low and underestimates $$$\mu$$$FA when IVF is high. SMT overestimates $$$\mu$$$MD in most cases. This stems from the fact the SMT does not include an isotropic compartment in its model and hence suffer from free-water contamination. SMT was reported to be inaccurate when the heterogeneity in a voxel is high (i.e., the voxel contains more than one compartment) [6].
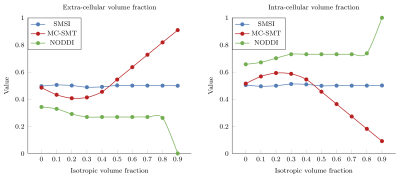
Intra- (ICVF) and extra-cellular volume fraction (ECVF): Fig. 2 shows the intra- and extra-cellular volume fraction estimated by MC-SMT, NODDI, and SMSI. SMSI yields results that are close to the ground truth (ECVF=ICVF=0.5). The MC-SMT tends to overestimate or underestimate the ICVF and the ECVF, depending on the IVF. Similar to SMT, MC-SMT does not include the isotropic compartment in its model. Hence, any signal contributed by the isotropic compartment will be explained by either the intra- or extra-cellular compartment in the model, biasing the results. Moreover, the tortuosity effect assumption of MC-SMT, i.e., $$$\lambda_{\bot}=(1-v_{ic})\lambda_{\parallel}$$$ does not always hold. NODDI, on the other hand, overestimates the ICVF and underestimates the ECVF.
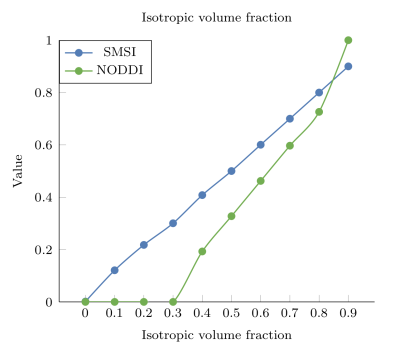
Isotropic volume fraction (IVF): Fig. 3 shows the isotropic volume fraction estimated by SMSI and NODDI versus the ground truth. The accuracy of SMSI in estimating the IVF is indicated by the almost straight line. NODDI, on the other hand, significantly underestimates the IVF when the actual IVF is low (<0.3) and introduces a moderate bias otherwise.
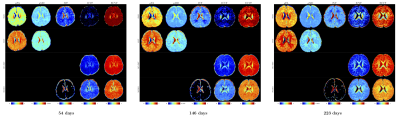
In-vivo results: Fig. 4 shows the results for the longitudinal in-vivo dataset of a baby subject. In agreement with the synthetic data results, we observed that:
- $$$u$$$FA and $$$u$$$MD: The SMT yields higher $$$u$$$FA and $$$u$$$MD in most of white matter and gray matter regions than SMSI. This is similar to observations drawn from the synthetic data that when IVF is moderately high (0.1<IVF<0.4) and when the microstructure is heterogeneous, SMT overestimates both $$$u$$$FA and $$$u$$$MD.
- ICVF and ECVF: NODDI and MC-SMT yields much higher ICVF and much lower ECVF than SMSI in most subcortical regions. This is consistent with the biases of NODDI and MC-SMT observed in the synthetic data experiments when the IVF is moderately high (0.1<IVF<0.4).
- IVF: NODDI yields zero IVFs in most subcortical regions, consistent with the observations in the synthetic data experiments.
Conclusions
We have demonstrated that the SMT, MC-SMT, and NODDI introduce biases in estimating anisotropy, diffusivity, and volume fractions of microstructral compartments. The bias is notable when the isotropic volume fraction is moderately high, which can happen in the baby data with greater water content. We have also shown that SMSI is robust to free water contamination and produces correct results in situations when SMT, MC-SMT, and NODDI fail.
Acknowledgements
This work was supported in part by NIH grants (MH117943, EB006733, AG041721, MH100217, NS093842, and 1U01MH110274) and the efforts of the UNC/UMN Baby Connectome Project Consortium.References
[1] Kaden, Enrico, Frithjof Kruggel, and Daniel C. Alexander. "Quantitative mapping of the per‐axon diffusion coefficients in brain white matter." Magnetic resonance in medicine 75.4 (2016): 1752-1763.
[2] Kaden, Enrico, et al. "Multi-compartment microscopic diffusion imaging." NeuroImage 139 (2016): 346-359.
[3] Daducci, Alessandro, et al. "Accelerated microstructure imaging via convex optimization (AMICO) from diffusion MRI data." NeuroImage 105 (2015): 32-44.
[4] Tiantian Xu, Geng Chen, Haiyong Wu, Weili. Lin, Dinggang Shen, Pew-Thian Yap.“Disentangling the Effects of Anisotropy and Orientation Dispersion Using Diffusion Spherical Mean Spectrum Imaging”, Joint Annual Meeting ISMRM-ESMRMB, Paris, France, 16-21 June 2018.
[5] Howell, Brittany R., et al. "The UNC/UMN baby connectome project (BCP): an overview of the study design and protocol development." NeuroImage (2018).
[6] Rafael Neto Henriques and Noam Shemesh. "Validity Regimes of the Spherical Mean Technique ", Joint Annual Meeting ISMRM-ESMRMB, Paris, France, 16-21 June 2018.
Figures