3563
MicroLearn: Framework for machine learning, reconstruction, optimization and microstructure modeling1Intelligent Systems Engineering, Indiana University Bloomington, Bloomington, IN, United States, 2University Medical Center Freiburg, Freiburg, Germany, 3The University of Minnesota Twin Cities, Minneapolis, MN, United States, 4Cardiff University, Cardiff, United Kingdom, 5Psychological and Brain Sciences, Indiana University Bloomington, Bloomington, IN, United States, 6Databricks, San Francisco, CA, United States
Synopsis
MicroLearn is a Machine Learning and Model Fitting framework that enables modular construction of multi-compartment microstructure models in crossings with fast and accurate parameter estimation.
Introduction
Recent research attempting to probe the tissue microstructure using diffusion MRI (dMRI), has led to a variety of model fitting and machine learning based approaches to estimate the underlying model parameters accurately and quickly 1. Both kinds of approaches construct multi-compartment models via combinations of intra- and extra- axonal spaces which are generally very hard to fit. MicroLearn aims at unifying these approaches with additional capabilities to interoperate between some state-of-the-art optimizers, machine learning methods and microstructure models. This framework is efficient in modeling regions of the brain with 0, 1, 2 and 3 fiber crossings alongside improved runtime performance and estimation accuracy. MicroLearn further helps underpin the mesostructure 2 (macro-level tissue organization) features necessary for reconstruction using Supervised Bayesian Learning of the tissue microstructure. The framework will be made available as a part of DIPY 3 with TensorFlow 4 integrations to easily switch between the training strategies, optimization methods and advanced regression algorithms. This novel framework includes transparent implementation of various models with a superquadrics-based visualization.Methods
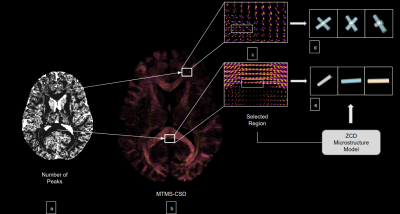
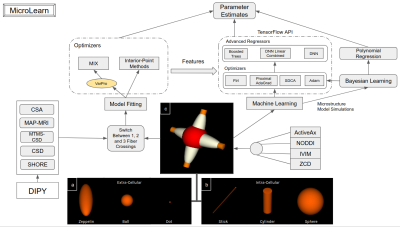
MicroLearn provides tools for estimating multi-compartment microstructure models by taking two different approaches: 1) Model Fitting based from Fiber Orientation Directions and 2) Machine Learning from Mesostructure. MicroLearn currently provides 4 different multi-compartment models, namely, ActiveAx 5, ZCD 6, NODDI 7, IVIM 8 in a modular manner without compromising on speed. These multi-compartment models can be fed into a model fitting module or into the machine learning module keeping the underlying microstructure model the same. For the Model Fitting paradigm, MicroLearn provides capabilities to switch between 0, 1, 2 and 3 fiber crossings for different tissues in specific regions of the brain. As shown in the figure, we can make use of the MTMS-CSD 9 (in combination with HMRF 10, SHORE 11, etc. from DIPY) to obtain the number of peaks in each voxel and fit the number of crossings accordingly (depicted in figure 2). In order to fit these complex biophysical models, MicroLearn provides tools for estimating parameters in manifold spaces using the Variable Projection 12 (VarPro) algorithm to find a global minima faster by constraining the non-linear parameters and separating them from the linear ones. This has been implemented in combination with differential evolution and convex optimizers to as a part of the MIX13 framework. Apart from this we also provide tools to perform interior point optimization using a different set of algorithms as an alternative to the MIX optimizer. In contrast to the above multi-stage fitting procedures, the machine learning paradigm provides tools to perform supervised Bayesian learning by extensively simulating and training signals2 from any of the above mentioned microstructure models. Along with a simple polynomial regression, MicroLearn provides advanced alternative training strategies via the TensorFlow API using Follow the regularized leader (Ftrl), Steepest Dual Coordinate Ascent (SDCA), Proximal AdaGrad and Adam optimizers for regressors such as Deep Neural Network (DNN), Boosted Trees and Linear Combined DNNs4. The learning approach is governed by a higher level mesoscopic structure to disentangle the meso- and micro- properties without having to use Fiber Orientation Distribution (FOD) as a prerequisite. This approach works with processed signal derivatives2 and extensive simulations of the underlying tissue microstructure model. As an added capability, we also provide means to train the microstructure models from the features obtained from the model fitting module (explained in detail in Figure 1).Results
This framework provides capabilities such as switching in and out from a machine learning to classical model fitting approach not currently available in any other toolbox. This implementation outperforms the estimation performances of NODDI, IVIM, ZCD and ActiveAx (by ~25X - 30X) with enhancements such as tools to fitting multiple fiber crossings. The experiments have been conducted on an intel i7 machine with no external GPUs required for computation. Contemporary toolboxes such as MDT 14, Dmipy 15, AMICO 16, and NODDI 7 implement microstructure toolboxes with modular and different approaches but do not provide flexible means to perform machine learning and direct model fitting at the same time.Discussion and Conclusion
MicroLearn provides computational capabilities from different parameter estimation paradigms with an end-goal of continually supporting more algorithms for transparent and collaborative growth of the microstructure domain. The framework will be available as a part of DIPY with efficient software design to help the community with a capability to adopt new models and approaches. Furthermore, it includes tutorials and experiments to compare different models and try different combinations with the Human Connectome Project (HCP) data.Acknowledgements
We would like to thank NIH R01EB027585 for supporting the PI and first author.References
[1] Ferizi et al. A Ranking of Diffusion MRI Compartment Models with In Vivo Human Brain Data. Magnetic Resonance in Medicine 72, 1785–1792; 2014.
[2] Reisert et. al. Disentangling micro from mesostructure by diffusion MRI: a bayesian approach NeuroImage, 147, 2017.
[3] Garyfallidis et. al. DIPY, a library for the analysis of diffusion MRI data, Frontiers in Neuroinformatics, vol. 8, p. 8, Frontiers, 2014.
[4] Abadi et al. Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv preprint arXiv:1603.04467, 2016.
[5] Dyrby et. al. Contrast and stability of the axon diameter index from microstructure imaging with diffusion MRI. Magnetic Resonance in Medicine 70, 711–721, 2013.
[6] Zhang et. al. Axon diameter mapping in crossing fibers with diffusion MRI. Medical image computing and computer-assisted intervention. MICCAI 14, 82–89, 2011.
[7] Zhang et. al. NODDI: Practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage 61, 1000–1016, 2012.
[8] Le Bihan et al. Separation of diffusion and perfusion in intravoxel incoherent motion MR imaging. Radiology 168.2, 1988.
[9] Jeurissen et. al. Multi-tissue constrained spherical deconvolution for improved analysis of multi-shell diffusion MRI data, NeuroImage, 103, 2014.
[10] Zhang et. al. Segmentation of Brain MR Images Through a Hidden Markov Random Field Model and the Expectation-Maximization Algorithm IEEE Transactions on Medical Imaging, 20(1): 45-56, 2001.
[11] Merlet et al., Continuous diffusion signal, EAP and ODF estimation via Compressive Sensing in diffusion MRI, Medical Image Analysis, 2013.
[12] Zhang, Y., et. al. Segmentation of Brain MR Images Through a Hidden Markov Random Field Model and the Expectation-Maximization Algorithm IEEE Transactions on Medical Imaging, 20(1): 45-56, 2001.
[13] Farooq et. al. Microstructure imaging of crossing (MIX) white matter fibers from diffusion MRI Sci. Rep., 6, 2016.
[14] Harms et. al. Robust and fast nonlinear optimization of diffusion MRI microstructure models, Neuroimage, 155, 2017.
[15] Fick et. al. Mipy: An Open-Source Framework to improve reproducibility in Brain Microstructure Imaging, OHBM, 2018.
[16] Daducci . et al. Accelerated Microstructure Imaging via Convex Optimization (AMICO) from diffusion MRI data. Neuroimage 105, 32–44, 2015
Figures