1184
Self-calibrating through-time spiral GRAPPA for flexible real-time imaging1Biomedical Engineering, Case Western Reserve University, Cleveland, OH, United States, 2Case Western Reserve University, Cleveland, OH, United States
Synopsis
A self-calibrating through-time spiral GRAPPA method is presented that uses the same undersampled scan for both the accelerated data and the calibration data. This method can be used to capture at least one heartbeat worth of data at 12 slice positions in less than one minute, in a free-breathing and ungated scan.
Introduction
Through-time non-Cartesian GRAPPA has been previously shown to yield cardiac images with high frame rates1. Unlike standard cine acquisitions, which require ECG gating and breathholding, non-Cartesian GRAPPA can be used to collect real-time images without gating during free breathing. Moreover, through-time GRAPPA is non-iterative and parallelizable, and data acquisition and reconstruction can be performed in real-time using GPUs2. However, one drawback of this technique is that a separate calibration dataset containing multiple repetitions of fully-sampled data must be acquired to calculate the non-Cartesian GRAPPA weights. In this work, a self-calibrated implementation of through-time GRAPPA is introduced, in which one undersampled scan is used for both the accelerated and calibration data. This work eliminates the need for a separate calibration scan, and enables flexible and rapid scanning. An initial application is shown for ejection fraction calculation, where a short-axis stack of 12 slices covering the left ventricle can be collected during free-breathing in less than one minute.Methods
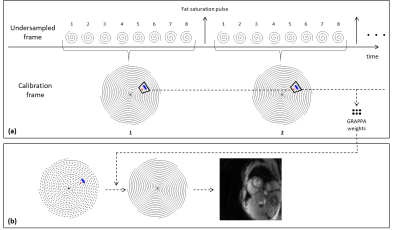
An 8-arm variable-density spiral trajectory3 was designed with the following parameters: TR/TE=10.56/0.84ms, FOV=300x300mm2, matrix size=128x128, slice thickness=8mm, in-plane resolution=2.34x2.34mm2. As shown in Fig.1, each undersampled frame consists of a single spiral arm, for an acceleration factor of 8. The eight spiral arms are collected consecutively, and each group of eight arms are merged together to create one fully-sampled calibration frame. The calibration and undersampled data can then be processed using the previously reported through-time spiral GRAPPA approach4.
The proposed self-calibrated through-time spiral GRAPPA method was tested on 8 healthy volunteers on a 3T clinical scanner (Skyra, Siemens Healthineers). Data were collected using a FLASH sequence with a flip angle of 15° and 30 receive coils from the spine and body arrays. Every 8 TRs, a 16.8ms fat saturation pulse was played. Twelve slices of data were collected with a slice gap of 0-20% to cover the left ventricle. At each slice, 320 undersampled frames with a temporal resolution of approximately 78fps (11ms/frame) were collected, with a total scan time of 4.05 seconds per slice. Note that this frame rate was calculated by distributing the time required for the saturation pulse over each TR. All spiral GRAPPA reconstructions were performed in the Gadgetron framework5. The number of coils was compressed to 12 before the GRAPPA reconstruction. A GRAPPA segment size of 8x1 (readout x projection arm) and a kernel size of 6x1 were used1.
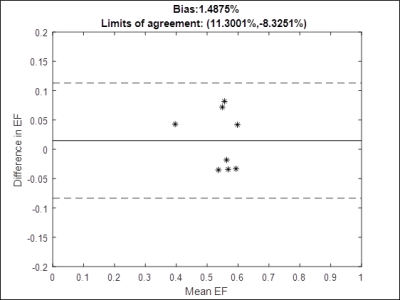
For comparison, standard breathheld and gated bSSFP cine scans were collected at the same slices. The parameters of the cine scan were: TR/TE=2.8/1.4ms, flip angle=40-60°, slice thickness=8mm, in-plane resolution=1.77x1.77mm2, bandwidth=965Hz/pixel, 25 cardiac phases. Ejection fractions (EF) were calculated by drawing ROIs over the blood pool in the left ventricle at diastole and systole at each slice. A Bland-Altman analysis was performed to assess the agreement between EFs derived from the standard cine and the proposed method.
Results
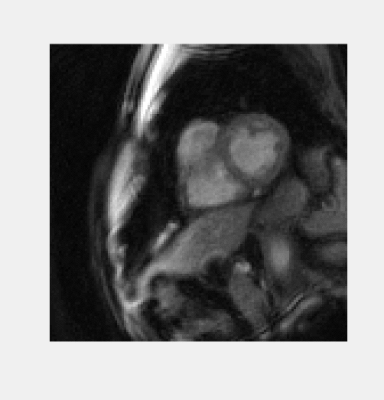
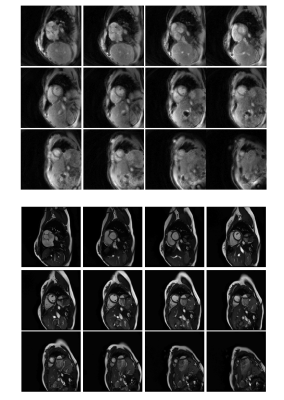
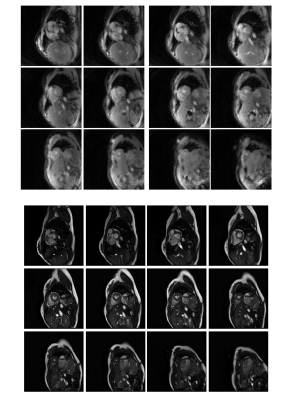
The total time to collect 320 undersampled frames in all 12 slices was approximately 50 seconds. An example video of images from one slice in one volunteer is shown in Fig.2. Fig.3,4 show comparisons of images from the stack of slices in diastole and systole using the proposed sequence and the standard cine. Fig.5 shows a Bland-Altman plot of the ejection fractions calculated using both sequences.Discussion
A self-calibrating through-time spiral GRAPPA method is proposed in which an undersampled scan is used for both accelerated and calibration data. One arm of data is acquired per accelerated frame, leading to a high overall frame rate and a short total scan time. Although the images from the proposed method appear to have more noise and poorer blood-myocardium contrast, the ejection fraction values are in acceptable agreement in this initial evaluation. One advantage of this technique is that the scan does not require breathholds or a regular heart rate. This may make it a good alternative for patients who cannot tolerate a cine scan, although further clinical evaluation is needed. This method may also be valuable in an interventional imaging setting, when it may be impractical to collect separate calibration data scans, and high frame rates are beneficial.
Future work may include using this approach with a bSSFP sequence for better blood-myocardium contrast, improving the spatial resolution, and including an off-resonance de-blurring step to compensate for the long readout time.
Conclusion
A self-calibrating through-time spiral GRAPPA method is presented for real-time cardiac imaging that eliminates the need for a separate calibration scan. This method can be used to capture at least one heartbeat of data at 12 slices in less than one minute, in a free-breathing and ungated scan.Acknowledgements
R01HL094557, NSF1563805, R01EB018108
References
1. Seiberlich N, Lee G, Ehses P, Duerk JL, Gilkeson R, Griswold M. Improved temporal resolution in cardiac imaging using through-time spiral GRAPPA. Magn. Reson. Med. 2011;66:1682–1688. doi: 10.1002/mrm.22952.
2. Franson D, Ahad J, Hamilton J, Lo W, Jiang Y, Chen Y, Seiberlich N. Real-time 3D cardiac MRI using through-time radial GRAPPA and GPU-enabled reconstruction pipelines in the Gadgetron framework. In: Proceedings of the 25th Annual Meeting of the International Society for Magnetic Resonance in Mediciine. Honolulu; 2017. p. 0448.
3. Hargreaves B. Variable-Density Spiral Design Functions. 2005. Available from: http://mrsrl.stanford.edu/~brian/vdspiral/
4. Hamilton JI, Wright KL, Griswold MA, Seiberlich N. Self-Calibrating Interleaved Reconstruction for Through-Time Non-Cartesian GRAPPA. In: Proceedings of the 13th Annual Meeting of the International Society for Magnetic Resonance in Medicine. Salt Lake City; 2013. p. 3836.
5. Hansen MS, Sørensen TS. Gadgetron: An open source framework for medical image reconstruction. Magn. Reson. Med. 2013;69:1768–1776. doi: 10.1002/mrm.24389.
Figures