1108
Fast 3D MR Fingerprinting with Pseudorandom Cartesian Sampling1Radiology, Case Western Reserve University, Cleveland, OH, United States, 2Siemens Healthcare GmbH, Erlangen, Germany, 3Friedrich‐Alexander‐Universität Erlangen‐Nürnberg, Erlangen, Germany, 4Biomedical Engineering, Case Western Reserve University, Cleveland, OH, United States, 5Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, MA, United States
Synopsis
A 3D Magnetic Resonance Fingerprinting method based on the pseudorandom Cartesian sampling was proposed to achieve isotropic high-resolution T1, T2 and off-resonance quantification. Compared to existing MRF methods based on non-Cartesian sampling patterns, the proposed method demonstrates a flexible sampling framework that is less susceptible to imperfections of the imaging gradients and off-resonance effects. It provides a step forward to a robust and high-resolution isotropic MRF method in brain, musculoskeletal and abdominal imaging.
Purpose
To develop a 3D Magnetic Resonance Fingerprinting (MRF) method based on pseudorandom Cartesian sampling with low-rank and subspace modeling to achieve high-resolution quantitative mapping.Introduction
MRF is a novel quantitative imaging technique to estimate multiple tissue properties simultaneously1. Most MRF methods have utilized non-Cartesian sampling patterns, such as spiral1,2, radial3, rosette4 and stack-of-spirals5,6 for 3D, as well as EPI trajectory7,8 to accelerate the acquisition. While these trajectories sample k-space more efficiently, they are susceptible to imperfections of the imaging gradients and off-resonance effects. In this work, an MRF method is proposed based on pseudorandom Cartesian sampling with temporal low-rank and subspace modeling9-12 to achieve high-resolution T1, T2 and off-resonance quantification without blurring artifacts due to off-resonance.Methods
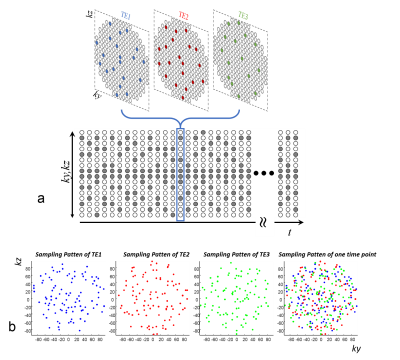
The proposed pseudorandom 3D Cartesian MRF-FISP2 acquisition used a Poisson-disc sampling pattern to acquire 4-D k-t space. A total of 500 time points were acquired with flip angles described previously2 and a constant TR of 5.68 ms. Each time point was highly undersampled with a pseudorandom sampling pattern as shown in Figure 1a. To encode off-resonance in this FISP prototype sequence, ky-kz readouts of each time point were segmented to acquire data at three different echo times13, while flip angle and TR were same. For example, the sampling pattern for one time point of a matrix size of 192 × 192 is shown in Figure 1b. The segment at each echo time was acquired with 96 readout lines, leading to an undersampling factor of R=384.
The reconstruction consisted of two steps. First, a low-rank reconstruction recovered a highly undersampled image of each time point, and then tissue properties were retrieved by taking the maximum of the inner product between the reconstructed images and the pre-calculated dictionary. The low-rank reconstruction was similar to existing low-rank reconstructions for MRF9-11 and T2-shuffling12. It formulated the reconstruction as
$$\min_{\alpha}\frac{1}{2}\parallel y + EU_{k}\alpha\parallel + \lambda\sum_a\parallel R_{r}(\alpha)\parallel _{*}$$
where $$$E$$$ is the encoding matrix that contains sampling masks and coil sensitives, $$$U_{k}$$$ is the subspace learned from the dictionary by using singular-value decomposition (SVD)14, $$$\alpha = U_k^H$$$ represents the compressed low-rank images, and $$$\sum_a\parallel R_{r}(\alpha)\parallel _{*}$$$ is a local low-rank regularization on low-rank images with block size $$$r$$$ of 8. The reconstruction was solved by FISTA15 with $$$\lambda$$$ = 0.01 of the maximum intensity of the low-rank images. It was implemented in Matlab using the BART toolbox16.
A dictionary containing a representative subset of potential signals was calculated using a Bloch equation simulation. It has 409,460 entries with T1 of 10 – 3000 ms, T2 of 1 – 800 ms, and off-resonance of -500 – 500 Hz. The dictionary was compressed into the temporal subspace by taking the first $$$k$$$ = 16 singular vectors of SVD.
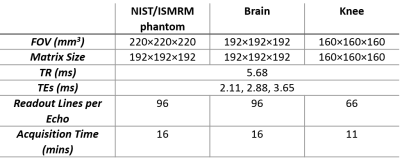
All experiments were performed on Siemens 3 T scanners (MAGNETOM Skyra and Prisma, Siemens Healthcare, Erlangen, Germany) with a 20-channel head receiver array for the phantom and the brain scans and a 15-channel TX/RX knee coil for the knee. Figure 2 shows the acquisition parameters for each experiment.
Results
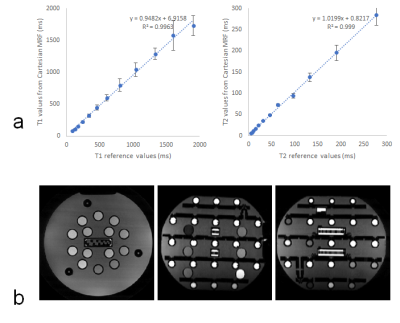
Figure 3a shows correlation curves that compare T1 and T2 values derived from the proposed method to the values reported by the NIST17. It demonstrates that the proposed MRF measurements are in good agreement with reference values. Figure 3b shows the images from the low-rank reconstruction corresponding to the first singular value from three orthogonal planes of the proposed method.
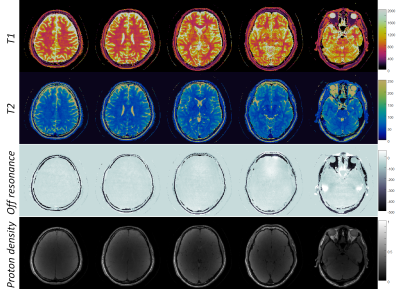
Figure 4 shows in vivo T1, T2, off-resonance and proton density maps in the brain, acquired using the proposed method. T1 and T2 values are 774±55.4 ms and 45±4.1 ms in white matter, 1236.7±96.9 ms and 70±9.1 ms in gray matter, which are in the range of previous reported values from 3D spiral MRF-FISP method5 .
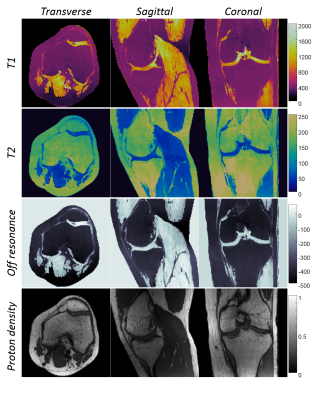
Figure 5 shows in vivo T1, T2, off-resonance and proton density maps of the knee. T1 and T2 values are 350±10 ms and 143±5.8 ms in bone marrow fat, 1230±61 ms and 37±7.1 ms in cartilage, and 1370±61 ms and 40±4.5 ms in muscle. They are in good agreements with previous reported values18.
Discussion and Conclusion
Compared to non-Cartesian MRF, the proposed method provides a flexible acquisition scheme based on a Cartesian acquisition. This Cartesian readout is much shorter than the previously used spiral readouts, which reduces the blurring artifacts that are encountered due to off-resonance effects. With variable echo times, this method encodes the off-resonance in the FISP readout, which has the potential to achieve chemical shift encoding for water-fat separation and to measure T2*. It provides a step forward to a robust and high resolution isotropic musculoskeletal MRF, and could be important in other applications such as abdominal imaging.Acknowledgements
This work was supported by Siemens Healthcare and NIH grants 1R01EB016728-01A1, 5R01EB017219-02, R01HL094557, and R01DK098503.References
1. Ma D, Gulani V, Seiberlich N, Liu K, Sunshine JL, Duerk JL, Griswold MA. Magnetic resonance fingerprinting. Nature 2013;495(7440):187-192.
2. Jiang Y, Ma D, Seiberlich N, Gulani V, Griswold MA. MR fingerprinting using fast imaging with steady state precession (FISP) with spiral readout. Magnetic resonance in medicine 2015;74(6):1621-1631.
3. Cloos, M.; Knoll F.; Zhao T.; Wiggins G.; Bruno M.; Block KT.; Sodickson DK. Multiparametric Imaging with Heterogeneous Radiofrequency Fields. Nature communications 2016; 7:12445.
4. Liu Y, Hamilton J, Griswold M, Seiberlich N. Simultaneous Quantification of T1 , T2 , and Off-resonance Using FISP-MRF with a Rosette Trajectory and Readout Segmentation. Proc. Intl. Soc. Mag. Reson. Med. 26th 2018;8246.
5. Ma D, Jiang Y, Chen Y, McGivney D, Mehta B, Gulani V, Griswold M. Fast 3D magnetic resonance fingerprinting for a whole-brain coverage. Magnetic resonance in medicine 2017;DOI:10.1002/mrm.26888.
6. Liaoc C, Bilgic B, Manhard M, Zhao B, Cao X, Zhong J, Wald L, Setsompop K. 3D MR fingerprinting with accelerated stack-of-spirals and hybrid sliding-window and GRAPPA reconstruction. NeuroImage 2017;162:13-22.
7. Rieger, B. , Zimmer, F. , Zapp, J. , Weingärtner, S. and Schad, L. R. (2017), Magnetic resonance fingerprinting using echo‐planar imaging: Joint quantification of T1 and T2* relaxation times. Magn. Reson. Med, 78: 1724-1733. doi:10.1002/mrm.26561
8. Cohen O, Zhu B, Rosen MS. MR fingerprinting Deep RecOnstruction NEtwork (DRONE). Magn Reson Med. 2018;80:885–894. https://doi.org/10.1002/mrm.27198
9. Zhao, B., Setsompop, K., Adalsteinsson, E. , Gagoski, B. , Ye, H. , Ma, D. , Jiang, Y. , Ellen Grant, P. , Griswold, M. A. and Wald, L. L., Improved magnetic resonance fingerprinting reconstruction with low‐rank and subspace modeling. Magn. Reson. Med., 2018;79: 933-942.
10. Assländer J, Cloos MA, Knoll F, Sodickson DK, Hennig J, Lattanzi R. Low rank alternating direction method of multipliers reconstruction for MR fingerprinting. Magn Reson Med. 2018;79(1):83-9
11. Hamilton, J. I., Jiang, Y. , Ma, D.,Chen, Y., Pawha, S., Lo, W. , Batesole J., Griswold, M. and Seiberlich, N. Low Rank Compressed Sensing Reconstruction for More Precise Cardiac MRF Measurement. ISMRM 2017 #554
12. Tamir, J. I., Uecker, M. , Chen, W. , Lai, P. , Alley, M. T., Vasanawala, S. S. and Lustig, M., T2 shuffling: Sharp, multicontrast, volumetric fast spin‐echo imaging. Magn. Reson. Med.,2017; 77: 180-195.
13. Pineda, A. R., Reeder, S. B., Wen, Z. and Pelc, N. J. (2005), Cramér–Rao bounds for three‐point decomposition of water and fat. Magn. Reson. Med., 54: 625-635. doi:10.1002/mrm.20623
14. McGivney, D. F., Pierre, E., Ma, D., Jiang, Y., Saybasili, H., Gulani, V., & Griswold, M. A. (2014). SVD compression for magnetic resonance fingerprinting in the time domain. IEEE transactions on medical imaging, 33(12), 2311-22.
15. A. Beck and M. Teboulle, A Fast Iterative Shrinkage-Thresholding Algorithm for Linear Inverse Problems. SIAM J Imaging Sci 2009, 2(1):183–202.
16. M. Uecker and J. I. Tamir. BART: version 0.4.02. Nov. 2017.
17. Keenan K, Stupic K, Boss M, et al. Multi‐site, multi‐vendor comparison of t1 measurement using ISMRM/NIST system phantom. In Proceedings of the 24th Annual Meeting of ISMRM, Singapore, 2016. Abstract 3290.
18. Gold GE, Han E, Stainsby J, Wright G, Brittain J, and Beaulieu C. Musculoskeletal MRI at 3.0 T: Relaxation Times and Image Contrast. American Journal of Roentgenology 2004 183:2, 343-351
Figures