0806
3D Magnetic Resonance Fingerprinting with Quadratic RF Phase1Radiology, Case Western Reserve University, Cleveland, OH, United States, 2Biomedical Engineering, Case Western Reserve University, Cleveland, OH, United States
Synopsis
Magnetic Resonance Fingerprinting with quadratic RF phase (qRF-MRF) has previously been reported for simultaneous quantitative mapping of T1, T2 and T2* relaxation times and off resonance for 2D acquisitions. Translation of qRF-MRF to 3D bears practical limitations for reconstruction and dictionary matching due to increase in data and dictionary sizes. Here, randomized SVD based time compression and reduction in dictionary size with quadratic fit are combined to overcome prohibitively large datasets and long reconstruction times of 3D qRF-MRF. Whole brain 3D qRF-MRF can be acquired in 5 minutes and is compared to 2D qRF-MRF and 3D FISP.
Introduction
Magnetic Resonance Fingerprinting1 maps multiple tissue properties and system parameters simultaneously. Recently, MRF with quadratic RF phase2 (qRF-MRF) method was developed to map off resonance (ΔB0), T1, T2, and T2* and validated in 2D. For qRF-MRF, on-resonance frequency is varied with a quadratic RF phase making the sequence sensitive to all possible frequencies available in the imaging object. Extending qRF-MRF method to 3D presents practical challenges with increases in data size and dictionary size with the additional two tissue property dimensions. In this work, the first implementation of 3D qRF-MRF was made possible by compression in time with randomized SVD3 (rSVD) and compression of the tissue property dimension with quadratic fit4. Fully matched whole brain maps of T1, T2, ΔB0 and T2* are obtained in as short as 5 minutes and are compared to standard MRF methods.Methods
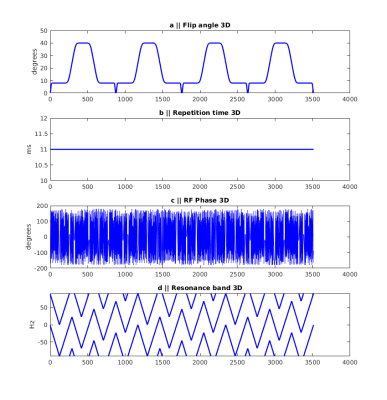
Data were collected with IRB approval and prior written consent. For the 3D qRF-MRF sequence, a 3D FISP sequence5,6 (300x300x144 mm3 FOV, 1.2x1.2x3 mm3 image resolution in ~10 mins; and 300x300x120 mm3 FOV, 1.2x1.2x5 mm3 image resolution in ~5 mins) was modified by removing the dephasing gradient before the RF pulse and varying the RF phase over time with a quadratic function. The phase change between consecutive RF pulses is small enough to avoid spoiling of MR signal and thus stay in TrueFISP regime. Sequence parameters and on resonance frequency variation over time are illustrated in Figure 1. Prior to reconstruction, both data and the target dictionary were compressed to rank 200 from 3516 time points. The truncation matrix was calculated with rSVD of a coarse dictionary with a small pool of tissue property and parameter values that spans the whole space. The qRF-MRF dictionary is formed by first simulating a dictionary with a range of T1, T2 and ΔB0 and then convolving it with a range of Γ width Lorentzian distributions. The Γ property is assumed to describe the frequency dispersion in a voxel and is used to calculate T2* afterwards2. Reconstructed signal evolutions were matched to the coarse dictionary to obtain the coarse tissue property maps. High resolution (in the tissue property dimension) was recovered with interpolation with a quadratic fit method. It is known that inner product values around the dictionary entry with the highest inner product form a quadratic curve. Considering the neighboring entries in four tissue property dimensions (81 combinations in total) a quadratic function is fit and the maximum along the fitted curve is taken as the new match. 3D qRF-MRF results are compared to single slice 2D qRF-MRF and matched FOV 3D FISP data.Results
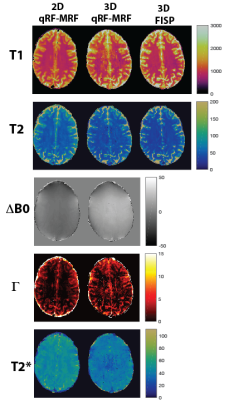
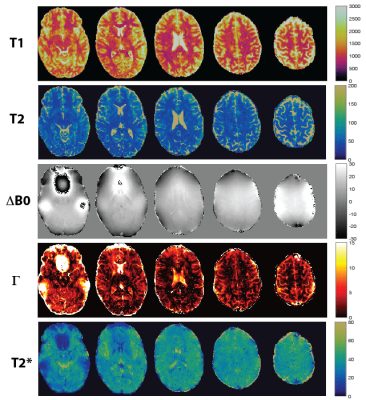
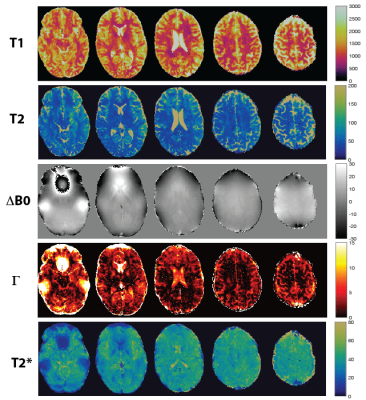
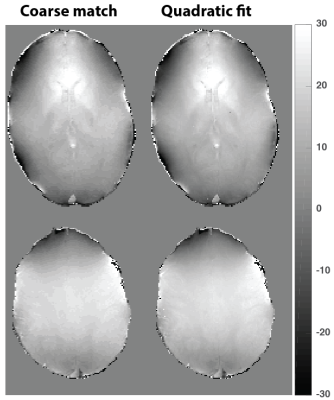
T1 and T2 maps of 2D qRF-MRF, 3D qRF-MRF and 3D FISP are compared in Figure 2. 3D qRF-MRF matches the T2 and T1 contrast of 3D FISP better than 2D qRF-MRF. In Figure 3 and 4, representative slices from the 3D qRF-MRF scan with 3mm and 5mm slices are plotted, respectively. Figure 5 illustrates the effect of quadratic fit on the coarse dictionary match for ΔB0.Discussion
3D qRF-MRF quantitative maps have partial blurring at the center of the image which are presumably from non-optimal coil geometry and possible B1 effects which needs to be accounted for. 3D FISP reconstruction can be reduced to as low as rank 25 with SVD whereas 3D qRF-MRF requires more with the increased variance in the tissue dimension due to the additional two dimensions (ΔB0 and Γ) in the dictionary. It was verified experimentally that ranks higher than 200 do not offer improvement in image quality but only come with additional overhead for image reconstruction and dictionary matching. Overestimation of T2 and underestimation of Γ for 2D qRF-MRF with respect to 3D qRF-MRF results in relatively longer T2* values. With the goal of isotropic resolution in a clinically feasible scan time, future work will focus on mitigating the central blurring problem and accelerating the acquisition with shortening of repeated flip angle and RF phase blocks.Conclusion
First implementation of 3D qRF-MRF for simultaneous mapping of T1, T2, off resonance and T2* is compared with its 2D counterpart and 3D FISP. Limitations in data and dictionary size are averted with compression in time with randomized SVD for and in the tissue dimension with the quadratic fit method. High resolution 3D qRF-MRF maps enable to go beyond traditional mapping and opens the door for advanced analyses with the novel and rich information provided in ΔB0 and T2* maps in 3D.Acknowledgements
The authors would like to acknowledge funding from Siemens Healthcare and NIH grants 1R01EB016728-01A1, 5R01EB017219-02 and R01 EB23704.References
1. Ma D, Gulani V, Seiberlich N, et al. Magnetic resonance fingerprinting. Nature 2013;495: 187–192.
2. Wang C, Coppo S, Mehta B, et al. Magnetic resonance fingerprinting with quadratic RF phase for measurement of T2* simultaneously with δf ,T1, and T2. Magn Reson Med, 2018, https://doi.org/10.1002/mrm.27543
3. Yang M, Ma D, Jiang Y, et al. Low rank approximation methods for MR fingerprinting with large scale dictionaries. Magn Reson Med 2018 79(4):2392-2400.
4. McGivney D, Boyacioglu R, Jiang Y, et al. Towards Continuous Dictionary Resolution in MR Fingerprinting using a Quadratic Inner Product Model. Submitted as an abstract, 27th annual ISMRM, Montreal (2019).
5. Ma D, Pierre E, McGivney D, et al. Applications of Low Rank Modeling to Fast 3D Magnetic Resonance Fingerprinting. ISMRM Proceedings 2017; p129.
6. Ma D, Jiang Y, Chen Y, et al. Fast 3D magnetic resonance fingerprinting for a whole-brain coverage. Magn Reson Med 2018, 79(4):2190-2197
Figures