0648
X-ray microcomputed tomography as a natively isotropic, nondestructive, 3D validation dataset for diffusion MRI1Department of Radiology, The University of Chicago, Chicago, IL, United States, 2Department of Neurobiology, The University of Chicago, Chicago, IL, United States, 3Argonne National Lab, Lemont, IL, United States
Synopsis
In this work, we present the use of synchotron x-ray microcomputed tomography (microCT) as a validation dataset for diffusion tensor imaging (DTI). DTI data were acquired of a post-mortem mouse brain. After metal staining, synchrotron microCT data of the sample were acquired, with 1.2 μm isotropic resolution across the whole brain. Orientation distribution functions were calculated from the microCT data using structure tensor analysis, and tractography was performed on the anterior commissure tract. Comparisons with tractography results from the diffusion MRI data show good agreement.
Introduction
Diffusion Tensor Imaging (DTI) is a powerful, non-invasive tool for characterizing neurological tissue microstructure on a macroscopic scale and is widely used in both research and clinical settings1,2. New methods of reconstructing orientation distribution functions (ODFs) from DTI data are rapidly being developed, each seeking to identify and model the distribution of distinct, sub-voxel axon fiber populations3.
Efforts to validate both ODF reconstruction methods and tractography results have typically relied on serial optical histology4-8. This requires the labor-intensive process of physically sectioning, staining, and optically scanning hundreds of slices of the tissue of interest. The slices are necessarily at least 10-20 times thicker than the achievable in-plane resolution (~5000 nm vs. ~250 nm), yielding non-isotropic volumetric reconstructions; distortions introduced by sectioning further limit the ability to align the slices and extract faithful information on the 3D orientation of fiber populations.
We are demonstrating the use of synchrotron micro computed tomography (microCT) as an alternative validation dataset. MicroCT allows for isotropic, nondestructive, 3D imaging of whole mouse brain specimens at sub-micron resolution, with the potential ability to resolve every axon in the brain.
Methods
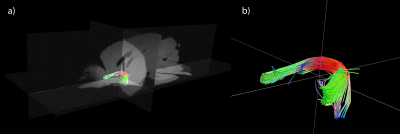
MRI data were acquired of a perfusion fixed control mouse brain. The brain was imaged with a Bruker 9.4 T magnet, using a 3D diffusion-weighted SE sequence at 150 μm isotropic resolution covering 30 directions and a b-value of 3000 s/mm2. The FSL tool DTIFIT was used to estimate the principal diffusion directions and other tensor parameters. Deterministic tractography was performed with Trackvis to visualize the anterior commissure.
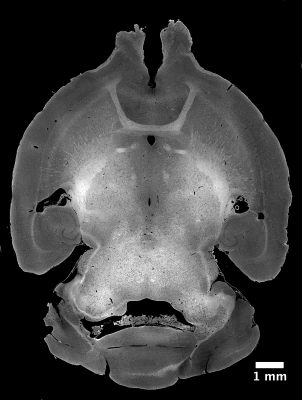
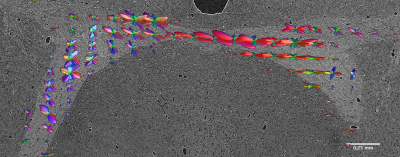
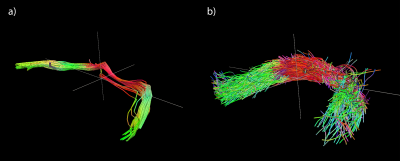
After MRI scanning, the same specimen was stained with uranyl acetate, osmium tetroxide and lead citrate in preparation for microCT imaging. Data were acquired at beamline 32-ID of the Advanced Photon Source at Argonne National Lab. The volume was reconstructed at 1.2 μm isotropic resolution using a mosaic sinogram-stitching method9. Voxel-wise local fiber orientation estimates were calculated from the microCT data using structure tensor analysis4,10. These estimates were binned across ROIs the size of single MRI voxels and represented both with a tensor model, and as orientation distribution functions expanded on a basis of real spherical harmonic functions up to a maximum degree of 16. Deterministic tractography was performed with Trackvis on the tensor model, and probabilistic tractography was performed with MRTrix on the ODF model to visualize the anterior commissure tract in the microCT data.
Results
Figure 1 shows the results of deterministic tractography calculated with Trackvis using a tensor model from the MRI data. Figure 2 shows a representative, full coronal slice of reconstructed microCT data, while Figure 3 displays visualizations of the ODFs calculated with structure tensor analysis in an ROI containing the anterior commissure. Figure 4 shows the results of both deterministic tractography from the tensor representation and probabilistic tractography from the ODF representation of the microCT data.Discussion
The tractography results from the microCT data show good qualitative agreement with those from MRI, verifying the accuracy of the local orientation estimates. Both determinsitic and probabilistic methods were able to recover the general shape and orientation of the anterior commissure. The probabilistic tractogram shows more variance in the paths of the streamlines. Future segmentation of the individual axons in the microCT data will allow us to quantify the true dispersion within the tract.Conclusions
These preliminary results demonstrate the feasibility and advantage of using microCT as a DTI validation dataset. The natively isotropic resolution of the microCT data allows for the direction calculation of local orientation estimates without the need for deconvolution and other pre-processing steps necessary for correcting anisotropic distortions in histology data. Additionally, microCT is a non-destructive, whole-brain imaging modality, which greatly simplifies the image registration process needed for quantitative validation.
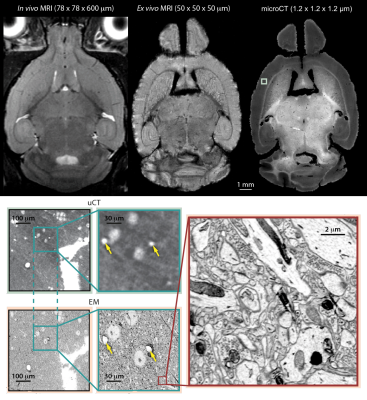
The potential to segment individual axons across the whole brain without the need for neural tracers will provide an unparalleled ground-truth dataset for MRI tractography validation. The stains used in preparation for microCT imaging are also used for contrast in serial electron microscopy. Accordingly, this work is part of an effort to develop a full validation pipeline for DTI, using imaging techniques with resolution spanning six orders of magnitude (mm to nm) applied in series to the same tissue. This pipeline is illustrated in Figure 5.
Acknowledgements
This work was partly funded by a Argonne National Laboratory - University of Chicago collaborative grant.
This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
References
[1] P. J. Basser, J. Mattiello, and D. LeBihan, “MR diffusion tensor spectroscopy and imaging.,” Bio- physical journal, vol. 66, pp. 259–67, jan 1994.
[2] W.-S. Tae, H. Byung-Joo, P. Sung-bom, S.-H. Kang, and B.-J. Kim, “Current Clinical Applications of Diffusion-Tensor Imaging,” Journal of Clinical Neurology, vol. 14, no. 2, pp. 129–140, 2018.
[3] M. Descoteaux, “High Angular Resolution Diffusion Imaging (HARDI),” Wiley Encyclopedia of Elec- trical and Electronics Engineering, pp. 1–25, 2015.
[4] M. D. Budde and J. A. Frank, “Examining brain microstructure using structure tensor analysis of histological sections,” NeuroImage, vol. 63, no. 1, pp. 1–10, 2012.
[5] C. Mitter, A. Jakab, P. C. Brugger, G. Ricken, G. M. Gruber, D. Bettelheim, A. Scharrer, G. Langs, J. A. Hainfellner, D. Prayer, and G. Kasprian, “Validation of In utero Tractography of Human Fetal Commissural and Internal Capsule Fibers with Histological Structure Tensor Analysis,” Frontiers in Neuroanatomy, vol. 9, no. December, pp. 1–15, 2015.
[6] A. Seehaus, A. Roebroeck, M. Bastiani, L. Fonseca, H. Bratzke, N. Lori, A. Vilanova, R. Goebel, and R. Galuske, “Histological validation of high-resolution DTI in human post mortem tissue,” Frontiers in Neuroanatomy, vol. 9, no. July, pp. 1–12, 2015.
[7] A. R. Khan, A. Cornea, L. A. Leigland, S. G. Kohama, S. N. Jespersen, and C. D. Kroenke, “3D structure tensor analysis of light microscopy data for validating diffusion MRI,” NeuroImage, vol. 111, pp. 192–203, 2015.
]8] K. G. Schilling, V. Janve, Y. Gao, I. Stepniewska, B. A. Landman, and A. W. Anderson, “Histological validation of diffusion MRI fiber orientation distributions and dispersion,” NeuroImage, vol. 165, pp. 200–221, jan 2018.
[9] R. F. C. Vescovi, M. B. Cardoso, and E. X. Miqueles, “Radiography registration for mosaic tomog- raphy,” Journal of Synchrotron Radiation, vol. 24, no. 3, pp. 686–694, 2017.
[10] J. Bigun and G. H. Granlund, “Optimal orientation detection of linear symmetry,” pp. 433–438, IEEE, 1987.
Figures