Tractography
1The Florey Institute of Neuroscience and Mental Health, Melbourne, Australia
Synopsis
In this introductory course on diffusion MRI based tractography, I will introduce the basics of tractography. I will by no means provide a comprehensive overview or review of all possible techniques and literature, but rather focus on the most essential basics of this area and a selection of its applications. As a result of attending this course, participants should be empowered to participate in the ongoing discussions about the strengths and limitations of tractography, as well as make informed and critical decisions on the use of tractography in their own work.
Audience and purpose
This course is intended for beginner and intermediate level attendees, interested in being introduced to some of the basics of diffusion MRI guided tractography. For a more comprehensive review of techniques, I would refer attendees to a recent review paper by Jeurissen et al.[1], which is also a good starting point for attendees wanting to prepare themselves for the course, or looking for more sources afterwards.The premise of tractography
Diffusion MRI is a unique tool that allows us to investigate the microstructure of white matter (WM) orders of magnitude smaller than the voxel size, in vivo, non-invasively and without ionising radiation. One of the properties of WM that can be accessed locally, is information on the local orientation of coherent bundles of axons (Fig.1–2, top panels). This information provides the basis for fibre tractography, which aims to reconstruct longer range bundles which fit the local orientational information well. More advanced strategies also try to fit other local properties, including variants of the local density of axons.Flavours of tractography
While the current range of tractography approaches is very broad, ranging from relatively generic to highly specialised techniques, several typical classifications have been used traditionally to categorise these approaches. These classifications include (yet are far from limited to):
- What kind of diffusion MRI model or information is being used to inform tractography: initially, the diffusion tensor model was very popular for this purpose (Fig.1), but it was later discovered to be relatively limited in its ability to describe the local orientational distribution of axons well. So called "crossing fibre" configurations, where essentially more than a single unique orientation is required to represent the distribution of axons within a voxel well, have been found to be quite prevalent across the white matter. More advanced models are able to overcome some of those limitations; an example is provided in Fig.2.
- Local ("streamline") versus global tractography: traditionally, local tractography techniques reconstruct streamlines by seeding from a (collection of) point(s) in the WM, and advancing in a step-wise fashion through the WM, making use of the local orientation information at each step (all figures feature results from streamline tractography techniques). Global tractography techniques aim to fit tracks directly to the data, while potentially interacting with each other to represent the data well. Several global tractography techniques do this by letting tracks arise from (initially) individual segments, which interact with each other via certain priors to align and connect the segments, as well as with the local data. Finally, there are also techniques that sit somewhere along the spectrum from local to global tractography; typically using some wider neighbourhood information to limit the step-wise accumulation of errors which poses a challenge to (local) streamline tractography.
- Within the category of local streamline tractography, both deterministic and probabilistic algorithms exist: while deterministic algorithms trace only a single unique path from a given seed point (example in Fig.1), probabilistic tractography allows for a whole distribution of streamlines emanating from each single seed point (example in Fig.2). Different probabilistic tractography algorithms use different definitions of the local probability distribution that is sampled from at each step along the streamline. Some of these take into account uncertainty due to noise and/or artefacts in the data, while others focus on uncertainty due to the actual (continuous) orientation distribution of fibres, or both. Probabilistic algorithms are often regarded as less prone to false negatives, but potentially more at risk of producing larger numbers of false positive streamlines.
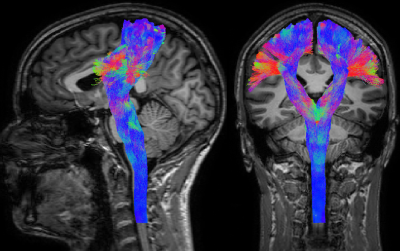
- Targeted versus whole-brain tractography: these are not so much categories of algorithms like the aforementioned ones, but rather different purposes for which any of those algorithms can be used. In whole-brain tractography, tracks are reconstructed for the entire volume of white matter, typically with the aim to discover which bundles exist and/or which areas of the brain they connect (examples in Fig.1–2). In targeted tractography, extra constraints are added, including areas which the tracks should visit (typically regions in which the end-points are expected to be located) and/or areas which the tracks are not allowed to visit. The aim here is to reconstruct a very specific (set of) known bundle(s), based on prior established anatomical information (example in Fig.3).
Finally, most (if not all) tractography algorithms are characterised by a set of (relatively ad hoc) parameters which control (constrain or regularise) further aspects of the reconstructed tracks, including step/segment size, curvature, length, coherence, etc...
Applications of tractography
Targeted tractography in particular provides an appealing semi-supervised segmentation mechanism for reconstruction of known white matter structures. While this can be useful for region of interest type of analyses, it has also grown to be regarded as a useful addition to the information used for pre-surgical planning. A typical example is shown in Fig.3: reconstruction of the corticospinal tract is often performed with the intention of avoiding damage to this structure during surgery, as it is crucial for motor and sensor functions (which are obviously important with respect to quality of life of patients).
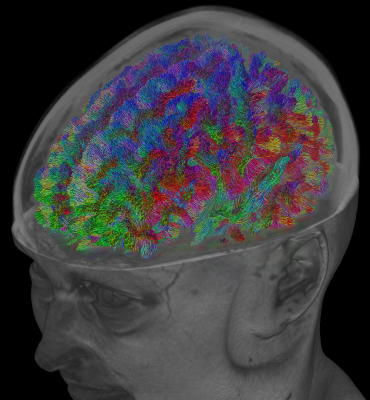
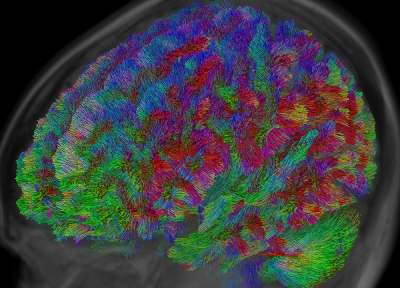
Whole-brain tractography has grown to be popular for the purpose of "connectomics": a set of analysis techniques which investigate all possible connections in the brain as a whole, and aim to characterise the resulting network. Essential in this context is the accuracy with which whole brain tractography can discover all bundles.
Problems and controversies
The accuracy of whole-brain tractography has recently come under scrutiny (again). While false negatives can be avoided relatively well nowadays, the extremely ill-posed nature of the whole-brain tractography problem renders it equally extremely vulnerable to false positives. A recent publication[2], based on results from an ISMRM 2015 tractography challenge, claims to provide data showing that even state-of-the-art tractography algorithms still produce a staggering number of false positive bundles. The publication[2] also doesn't avoid several strongly worded statements:
- "Tractograms contained more invalid than valid bundles."
- "[...] thick and dense bundles of plausible looking streamlines in locations where such streamlines did not actually exist."
- "This demonstrates the inability of current state-of-the-art tractography algorithms to control for false positives."
This is quite remarkable, given the long list of co-authors (who consequently endorse these statements) including researchers across the domain, combined with the long review process and several unambiguous warnings from reviewers[3] throughout this process. This provides an interesting insight (for beginners and experts alike) and public record of some of the currently ongoing debate about the applicability of these techniques; not unimportant, considering that whole-brain tractography is a fundamental basis that popular (structural) connectomics analysis techniques heavily rely upon.
Conclusion
Tractography exploits unique information available to us via diffusion MRI to reconstruct axon bundles in the WM up to some extent. Successful applications of targeted tractography include adding information to pre-surgical planning. Whole-brain tractography is extremely challenging, with major outstanding issues. This challenges the validity of analyses based on whole-brain tractography.Acknowledgements
Figures 1–5 were generated using MRtrix3 (http://www.mrtrix.org).References
[1] Jeurissen B, Descoteaux M, Mori S, Leemans A. Diffusion MRI fiber tractography of the brain. NMR Biomed. 2017, Sep 25. (doi: 10.1002/nbm.3785)
[2] Klaus H. Maier-Hein, Peter F. Neher, Jean-Christophe Houde, Marc-Alexandre Côté, Eleftherios Garyfallidis, Jidan Zhong, Maxime Chamberland, Fang-Cheng Yeh, Ying-Chia Lin, Qing Ji, Wilburn E. Reddick, John O. Glass, David Qixiang Chen, Yuanjing Feng, Chengfeng Gao, Ye Wu, Jieyan Ma, H. Renjie, Qiang Li, Carl-Fredrik Westin, Samuel Deslauriers-Gauthier, J. Omar Ocegueda González, Michael Paquette, Samuel St-Jean, Gabriel Girard, François Rheault, Jasmeen Sidhu, Chantal M. W. Tax, Fenghua Guo, Hamed Y. Mesri, Szabolcs Dávid, Martijn Froeling, Anneriet M. Heemskerk, Alexander Leemans, Arnaud Boré, Basile Pinsard, Christophe Bedetti, Matthieu Desrosiers, Simona Brambati, Julien Doyon, Alessia Sarica, Roberta Vasta, Antonio Cerasa, Aldo Quattrone, Jason Yeatman, Ali R. Khan, Wes Hodges, Simon Alexander, David Romascano, Muhamed Barakovic, Anna Auría, Oscar Esteban, Alia Lemkaddem, Jean-Philippe Thiran, H. Ertan Cetingul, Benjamin L. Odry, Boris Mailhe, Mariappan S. Nadar, Fabrizio Pizzagalli, Gautam Prasad, Julio E. Villalon-Reina, Justin Galvis, Paul M. Thompson, Francisco De Santiago Requejo, Pedro Luque Laguna, Luis Miguel Lacerda, Rachel Barrett, Flavio Dell’Acqua, Marco Catani, Laurent Petit, Emmanuel Caruyer, Alessandro Daducci, Tim B. Dyrby, Tim Holland-Letz, Claus C. Hilgetag, Bram Stieltjes & Maxime Descoteaux. The challenge of mapping the human connectome based on diffusion tractography. Nat Commun. 2017, Nov 7;8(1):1349. (doi: 10.1038/s41467-017-01285-x)
[3] The peer review file is publicly available as part of the electronic supplementary material, accessible via the online version of the published article at https://www.nature.com/articles/s41467-017-01285-x
Figures
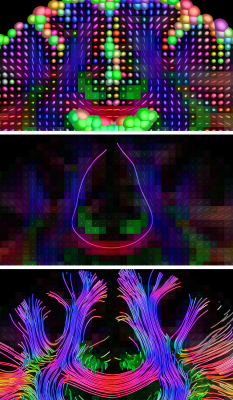
Fig.1: deterministic tractography guided by the first eigenvector ("main orientation") from the diffusion tensor model.
Top: the diffusion tensors in part of a coronal slice.
Middle: a single deterministic streamline seeded from the midbody of the corpus callosum, tracking eigenvectors.
Bottom: whole-brain result, seeded randomly across the white matter.
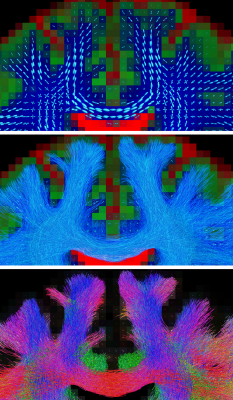
Fig.2: probabilistic tractography guided by fibre orientation distributions (FODs) from a multi-tissue model, accounting for crossing fibres and presence of non-WM tissues.
Top: the FODs and tissue compartments from the multi-tissue model.
Middle: probabilistic whole-brain tractography using all orientational information of the FODs (i.e., their entire shape; not just the peaks).
Bottom: same result, with traditional directional colouring, showcasing the mix of different orientations of streamlines in the crossing areas.