5644
Improved quantification of cerebral blood flow in gray matter and white matter using non-Fourier based reconstructionPhil Lee1,2, Peter Adany2, and In-Young Choi1,2,3
1Molecular & Integrative Physiology, University of Kansas Medical Center, Kansas City, KS, United States, 2Hoglund Brain Imaging Center, University of Kansas Medical Center, Kansas City, KS, United States, 3Neurology, University of Kansas Medical Center, Kansas City, KS, United States
Synopsis
Accurate quantification of CBF in GM and WM is challenging due to intrinsically low SNR and low spatial resolution of CBF maps. In this study we propose a new approach in quantifying CBF in GM and WM, based on spectral localization by imaging (SLIM), which provides accurate CBF values in compartments with complex shapes, e.g., GM and WM by incorporating high-resolution anatomical information into CBF reconstruction at reduced scan time.
Purpose
Cerebral blood flow (CBF) is commonly measured non-invasively using arterial spin labeling techniques [1]. Despite recent advancement of MR technology in CBF measurements, the spatial resolution of CBF maps is relatively poor due to the intrinsically low signal-to-noise ratio (SNR) of perfusion signals on the order of 1–2% of general MRI signals. Most CBF techniques cannot provide sufficient spatial resolution to accurately quatify CBF in gray matter (GM) and white matter (WM) separately because of the discrepancy between achievable spatial resolution of CBF measurement and required spatial resolution given the thickness of cortical GM in humans (1.5–3 mm), resulting in under- and over-estimation of CBF in GM and WM, respectively. Furthermore, significant differences of CBF between GM and WM (~2 folds) demand more stringent requirement of the spatial localization accuracy for CBF measurements. In this study we propose a new approach to quantify CBF in GM and WM based on spectral localization by imaging (SLIM) technique [2], namely SLIM-CBF, which incorporates high-resolution anatomical information into CBF reconstruction and provides accurate CBF values in compartments with complex shapes, e.g., GM and WM.Methods
Healthy subjects were studied on a 3 T (Skyra, Siemens) scanner with a 16 channel head receive coil. The CBF data were acquired using a pseudo-continuous arterial spin lableing technique (TE/TR=49/4300 ms, spatial resolution=2.5x2.5x3 mm3, slice thickness=3 mm, number of averages=9, label offset=9 cm) [3-4]. High resolution T1-weighted MRI was acquired using a magnetization-prepared rapid acquisition gradient echo (MPRAGE) sequence (TE/TR=4.4/2500 ms, TI=1100 ms, spatial resolution=1 mm3). MRI volumes were coregistered using coregistration tools in AFNI and FSL. Image segmentation of GM, WM, and CSF compartments were performed using automatic segmentation routines (FSL BET, SPM8). CBF maps were calculated using presaturated labeled and control images, and non-presaturated M0 images assuming the labeling efficiency of 0.68 [5]. GM and WM compartments were generated using GM and WM segmented maps and were sub-divided into three sub regions (frontal, fronto-parietal, and parietal). SLIM-CBF reconsruction was performed using coregistered high-resolution comparmental masks and CBF maps [2,6]. SLIM reconstruction involves generalized matrix inversion of SLIM constitution equation, sn = Gnm cm, where sn is the measured k-space signals, cm comparmental signals, Gnm the SLIM geometry matrix defined as ΣDm exp(-j 2π kn r), the Dm mth compartment, and kn the nth k-space encoding vector. The outcome of SLIM reconstuction is mean complartmental signals. Performance of SLIM-CBF reconstruction was tested using down-sampled single-slice CBF input maps in k-space with the matrix sizes of 8x8, 16x16, 32x32, 64x64 and 128x128 to test feasibility of reducing scan time. The effect of SNR on the estimated GM and WM CBF values was tested by successively increasing the number of averages of the CBF MRI data.Results and Discussion
The proposed SLIM-CBF reconstruction showed a consistent GM and WM CBF contrast at a variety of input CBF resolutions in constrast to severe blurring in the Fourier transform reconstruction (Fig. 1). The SLIM-CBF reconstruction showed more robust and accurate estimation of GM and WM CBF values than the conventional mask-based estimation, as demonstrated by larger differences of CBF values between GM and WM similar to known CBF differences between GM and WM even at very low spatial resolution of 32x32, corresponding to the pixel size of 10x10 mm2 (Fig. 2). The major advantage of the SLIM-CBF reconstruction is its capability to acquire accurate GM and WM CBF values using low resolution CBF MRI, which significantly accelerate the CBF data acquisition, e.g., reducing scan time by 16-folds in the case of reducing matrix size from 128x128 to 32x32 with the same SNR. In conclusion, the SLIM-CBF reconstruction is a promising strategy to accurately measure CBF values in compartments with complex shapes, where conventional approaches fail because of the limited spatial resolution due to intrinsically low SNR of CBF MRI.Acknowledgements
The Hoglund Brain Imaging Center is supported by the NIH (S10RR029577) and the Hoglund Family Foundation.References
- Detre et al., MRM (1992) 23:37-45.
- Hu et al., MRM (1988) 8:314-322.
- Dai et al., MRM (2008) 60:1448-1497.
- Fernandez-Seara et al., MRM (2008) 59:1467-1471.
- Wang et al., MRM (2003) 50:599-607.
- Adany et al., NeuroImage (2016) 134:355-364.
Figures
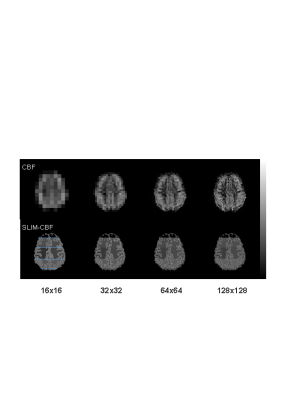
Figure 1. CBF maps and corresponding SLIM-CBF
maps at various spatial resolutions. Gray bar indicates CBF values of 0-100
ml/100g/min. Brain regions were divided three areas: frotal, fronto-parietal,
and parietal regions, divided by blue horizontal lines in the bottom left
image.
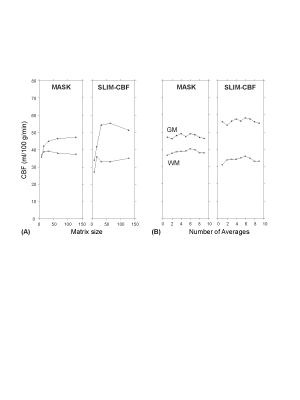
Figure 2. Comparisons of quantified CBF values
in GM and WM with mask-based method vs. SLIM-CBF method. (A) Relationship
between CBF values and spatial resolution. (B) Relationship between number of
averages and CBF values.