5640
Online Free-Breathing Liver Perfusion Imaging Using Parallel Computing and the Gadgetron Framework1Dept. of Biomedical Engineering, Case Western Reserve University, Cleveland, OH, United States, 2Dept. of Radiology, University Hospitals of Cleveland, Cleveland, OH, United States, 3School of Medicine, Case Western Reserve University, Cleveland, OH, United States
Synopsis
Accelerated image acquisition, advanced reconstruction and registration methods, and perfusion modeling have recently enabled free-breathing and quantitative 4D DCE MRI in the liver. However, the reconstruction, registration and model fitting steps are performed offline as they are time consuming; each data set takes more than a day to analyze. This makes quantitative 3D liver perfusion unsuitable for clinical deployment. We propose using a GPU-based Gadgetron framework for parallelized and near-immediate provision of all data, including a Spiral GRAPPA reconstruction, non-rigid image registration, and pharmacokinetic modeling using a dictionary-based approach. This approach reduces the reconstruction time from hours to minutes.
Purpose
While image reconstruction and processing techniques continue to advance, the required computational intensity has concomitantly increased. Modern reconstructions are often time consuming and hard to implement in the clinical setting. The goal of this work was to develop an online framework to provide free-breathing, high-spatiotemporal resolution DCE liver MRI data using a GPU-based online Spiral GRAPPA reconstruction in the Gadgetron framework, coupled with efficient parallelized online image registration to reduce respiratory motion and perfusion quantification to estimate pharmacokinetic properties.Method
All acquisitions were performed on a Siemens 3T scanner (Skyra, Siemens Healthineers, Erlangen, Germany). Free-breathing, accelerated stack-of-spirals data were acquired as described by Chen, et al2. A calibration scan was acquired, consisting of 3 fully sampled 3D volumes, and the data acquisition followed, with an acceleration factor of 6 (8/48 spiral arms). The following parameters were utilized: FOV 420mm; resolution 1.9x1.9x3 mm3; temporal resolution ~2s/volume; 100 frames, flip angle 10°.
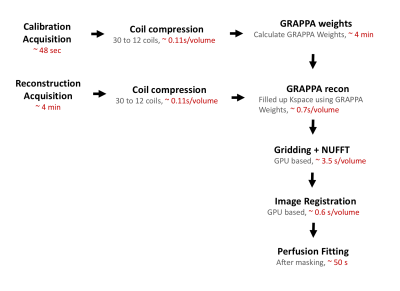
The Gadgetron reconstruction process was triggered when data acquisition was initiated. GRAPPA weights were first estimated using fully sampled data, and these weights were used to reconstruct the undersampled DCE MRI data. Both GRAPPA calibration and reconstruction were performed after coil compression to reduce the computational load and accelerate the reconstruction speed. GPU-based gridding and NUFFT as well as coil combination were performed afterwards to generate images. GPU and parallel computing also has been used to speed up iterative linear and non-linear registration. Dictionary based perfusion quantification to estimate pharmacokinetic properties was implemented; an approach similar to that in Magnetic Resonance Fingerprinting5. A dual-input single-compartment (DISC) model6 was used to generate a dictionary of all possible enhancement timecourses, and data from the registered images were matched to the dictionary by calculating the maximum correlation coefficient. From this dictionary index, the perfusion properties Arterial Fraction (AF), Distribution Volume (DV) and mean transit time (MTT) can be obtained. Finally, the reconstructed images along with three maps were sent back to scanner in DICOM format.
Data were processed in both Matlab and the proposed Gadgetron pipeline. A reconstruction server was used to perform this framework, which has an 8GB Nvidia GTX 1080 graphics card; an 10 core, 2.4GHz Intel Xeon E5-2630 v4 processor; and 64GB (8 x 8GB) of 2400MHz DDR4 RAM. All the code was constructed based on C++, CUDA and OpenCL.
Results
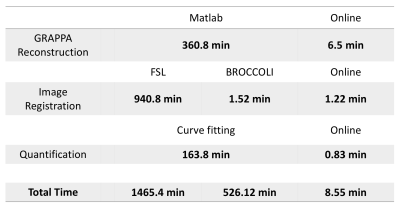
Figure 1 shows the Gadgetron framework flowchart and the computational time of each step. Table 1 shows the total time of each step. For example, in the GRAPPA reconstruction step, Matlab reconstruction took 3.6 minutes per volume for a total of 360.8 minutes2, while online reconstruction took 3.9 seconds per volume for a total of 6.5 minutes. Note this framework allows images to be sent back to the scanner before the whole scan is complete, the operator sees volumes continuously appearing, while scanning is ongoing.
Figure 2 shows Spiral GRAPPA images reconstructed using both offline Matlab and online Gadgetron implementations. No clear visual difference was observed, and Dice Similarity Coefficient showed the results are identical.
Figure 3 shows the effectiveness of using online image registration. Iterative linear and non-linear registration algorithms are applied to reduce respiratory motion. Parallel computing and GPU are involved to make the registration time cost 0.6s/volume, which is more efficient than BROCCOLI4 registration.
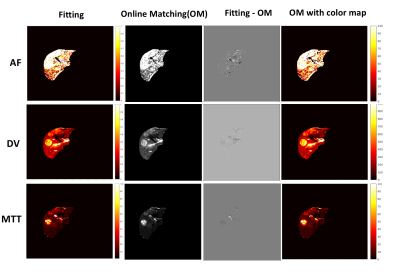
Figure 4 shows a comparison of quantification methods between traditional curve fitting and online dictionary matching. One slice of a 3D dataset from a patient with metastatic adenocarcinoma is shown. Dictionary matching estimation shows similar perfusion properties with curve fitting. For a 3D volume, curve fitting takes 2.73 hours, while online perfusion fitting using a dictionary approach only costs 50 seconds.
Conclusion and Discussion
In this work, an online reconstruction framework was developed for through-time Spiral GRAPPA reconstruction, image registration and perfusion fitting. The results show a dramatic time reduction in processing. The reconstruction, registration and quantification framework is developed on the open source Gadgetron platform. Thus, the entire pipeline or individual steps of this process could be easily deployed on multiple scanners or on different vendor platforms, which enables a broader clinical use. Future work will involve deployment of this framework at multiple sites. One current limitation is arterial inputs are manually selected and areas outside of the liver parenchyma are masked to improve efficiency. Automation of these steps will also be explored in the future.
Acknowledgements
The authors would like to acknowledge funding from 1R01DK098503 and Siemens Healthineers.References
1. Chen Y, et al. Invest Radiol, 2015; 50:367-375.
2. Hansen M, et al. MRM, 2013; 69.6: 1768-1776
3. Seiberlich N, et al. MRM. 2011; 66:1682–1688.
4. Ghodasara S, et al. ISMRM, 2016; 0161
5. Ma D, et al. Nature, 2013; 495:187-192
6. Materne R, et al. Clinical Science. 2000;99(6):517
Figures